 |
PDBsum entry 8f3d

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 0 more)
566 a.a.
(+ 0 more)
566 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 0 more)
678 a.a.
(+ 0 more)
678 a.a.
|
 |

|

|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
3-methylcrotonyl-coa carboxylase in filament, beta-subunit centered
|
|
Structure:
|
 |
3-methylcrotonyl-coa carboxylase beta-subunit. Chain: a, b, c, d, e, f. 3-methylcrotonyl-coa carboxylase alpha-subunit. Chain: h, i, j, k, l, m
|
|
Source:
|
 |
Leishmania tarentolae. Organism_taxid: 5689. Strain: tatii/uc. Strain: tatii/uc
|
|
Authors:
|
 |
J.J.Hu,J.K.J.Lee,Y.T.Liu,C.Yu,L.Huang,I.Afasizheva,R.Afasizhev, Z.H.Zhou
|
|
Key ref:
|
 |
J.J.Hu
et al.
(2023).
Discovery, Structure, And function of filamentous 3-Methylcrotonyl-Coa carboxylase..
Structure,
31,
100.
PubMed id:
|
 |
|
Date:
|
 |
|
09-Nov-22
|
Release date:
|
11-Jan-23
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F:
E.C.6.4.1.4
- methylcrotonoyl-CoA carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
3-methylbut-2-enoyl-CoA + hydrogencarbonate + ATP = 3-methyl-(2E)- glutaconyl-CoA + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
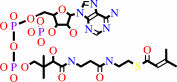 3-methylbut-2-enoyl-CoA
3-methylbut-2-enoyl-CoA
|
+
|
 hydrogencarbonate
hydrogencarbonate
|
+
|
 ATP
ATP
|
=
|
3-methyl-(2E)- glutaconyl-CoA
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Biotin
|
 |
 |
 |
 |
 |
Biotin
Bound ligand (Het Group name =
BTI)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
|

