
|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 275 a.a.
275 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 2 more)
256 a.a.
(+ 2 more)
256 a.a.
|
 |

|

|

|

|

|

|

|



 216 a.a.
216 a.a.
|
 |

|

|

|

|

|

|

|

 205 a.a.
205 a.a.
|
 |

|

|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Viral protein
|
 |
|
Title:
|
 |
Cryo-em structure of the mvv csc intasome at 4.5a resolution
|
|
Structure:
|
 |
Integrase. Chain: a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p. Fragment: unp residues 821-1101. Synonym: in. Engineered: yes. Vdna, non-transferred strand. Chain: q, s. Engineered: yes. Vdna, transferred strand.
|
|
Source:
|
 |
Visna/maedi virus ev1 kv1772. Organism_taxid: 36374. Strain: kv1772. Gene: pol. Expressed in: escherichia coli. Expression_system_taxid: 562. Synthetic: yes. Visna lentivirus (strain 1514). Organism_taxid: 11742.
|
|
Authors:
|
 |
A.Ballandras-Colas,D.Maskell,V.E.Pye,J.Locke,S.Swuec,A.Kotecha, A.Costa,P.Cherepanov
|
|
Key ref:
|
 |
A.Ballandras-Colas
et al.
(2017).
A supramolecular assembly mediates lentiviral DNA integration.
Science,
355,
93-95.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
28-Apr-22
|
Release date:
|
11-May-22
|
|
|
Supersedes:
|

|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P35956
(POL_VILVK) -
Gag-Pol polyprotein from Maedi visna virus (strain KV1772)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1506 a.a.
275 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P35956
(POL_VILVK) -
Gag-Pol polyprotein from Maedi visna virus (strain KV1772)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1506 a.a.
256 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.2.7.7.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.2.7.7.49
- RNA-directed Dna polymerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
DNA(n) + a 2'-deoxyribonucleoside 5'-triphosphate = DNA(n+1) + diphosphate
|
 |
 |
 |
 |
 |
 DNA(n)
DNA(n)
|
+
|
2'-deoxyribonucleoside 5'-triphosphate
|
=
|
DNA(n+1)
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.2.7.7.7
- DNA-directed Dna polymerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
DNA(n) + a 2'-deoxyribonucleoside 5'-triphosphate = DNA(n+1) + diphosphate
|
 |
 |
 |
 |
 |
 DNA(n)
DNA(n)
|
+
|
2'-deoxyribonucleoside 5'-triphosphate
|
=
|
DNA(n+1)
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.3.1.-.-
|
|
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.3.1.13.2
- exoribonuclease H.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Exonucleolytic cleavage to 5'-phosphomonoester oligonucleotides in both 5'- to 3'- and 3'- to 5'-directions.
|
 |
 |
 |
 |
 |
Enzyme class 6:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.3.1.26.4
- ribonuclease H.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Endonucleolytic cleavage to 5'-phosphomonoester.
|
 |
 |
 |
 |
 |
Enzyme class 7:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.3.4.23.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 8:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.3.6.1.23
- dUTP diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dUTP + H2O = dUMP + diphosphate + H+
|
 |
 |
 |
 |
 |
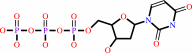 dUTP
dUTP
|
+
|
H2O
|
=
|
 dUMP
dUMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
355:93-95
(2017)
|
|
PubMed id:
|
|
|
|

|
| |
|
A supramolecular assembly mediates lentiviral DNA integration.
|
|
A.Ballandras-Colas,
D.P.Maskell,
E.Serrao,
J.Locke,
P.Swuec,
S.R.Jónsson,
A.Kotecha,
N.J.Cook,
V.E.Pye,
I.A.Taylor,
V.Andrésdóttir,
A.N.Engelman,
A.Costa,
P.Cherepanov.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |

| |

