 |
PDBsum entry 7jn4

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Plant protein
|
PDB id
|
|

|

|
7jn4
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|





 (+ 2 more)
427 a.a.
(+ 2 more)
427 a.a.
|
 |

|

|

|

|

|

|

|





 (+ 2 more)
138 a.a.
(+ 2 more)
138 a.a.
|
 |

|

|
|
|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Plant protein
|
 |
|
Title:
|
 |
Rubisco in the apo state
|
|
Structure:
|
 |
Ribulose bisphosphate carboxylase large chain. Chain: a, c, e, g, i, k, m, o. Synonym: rubisco large subunit. Ribulose bisphosphate carboxylase small chain 2, chloroplastic. Chain: b, d, f, h, j, l, n, p. Synonym: rubisco small subunit 2. Ec: 4.1.1.39
|
|
Source:
|
 |
Chlamydomonas reinhardtii. Organism_taxid: 3055. Organism_taxid: 3055
|
|
Authors:
|
 |
D.Matthies,M.C.Jonikas,S.He
|
|
Key ref:
|
 |
S.He
et al.
(2020).
The structural basis of Rubisco phase separation in the pyrenoid.
Nat Plants,
6,
1480-1490.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
03-Aug-20
|
Release date:
|
18-Nov-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D, E, F, G, H, I, J, K, L, M, N, O, P:
E.C.4.1.1.39
- ribulose-bisphosphate carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2 (2R)-3-phosphoglycerate + 2 H+ = D-ribulose 1,5-bisphosphate + CO2 + H2O
|
 |
 |
 |
 |
 |
2
×
(2R)-3-phosphoglycerate
|
+
|
2
×
H(+)
|
=
|
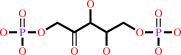 D-ribulose 1,5-bisphosphate
D-ribulose 1,5-bisphosphate
|
+
|
 CO2
CO2
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Plants
6:1480-1490
(2020)
|
|
PubMed id:
|
|
|
|

|
| |
|
The structural basis of Rubisco phase separation in the pyrenoid.
|
|
S.He,
H.T.Chou,
D.Matthies,
T.Wunder,
M.T.Meyer,
N.Atkinson,
A.Martinez-Sanchez,
P.D.Jeffrey,
S.A.Port,
W.Patena,
G.He,
V.K.Chen,
F.M.Hughson,
A.J.McCormick,
O.Mueller-Cajar,
B.D.Engel,
Z.Yu,
M.C.Jonikas.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Approximately one-third of global CO2 fixation occurs in a
phase-separated algal organelle called the pyrenoid. The existing data suggest
that the pyrenoid forms by the phase separation of the CO2-fixing
enzyme Rubisco with a linker protein; however, the molecular interactions
underlying this phase separation remain unknown. Here we present the structural
basis of the interactions between Rubisco and its intrinsically disordered
linker protein Essential Pyrenoid Component 1 (EPYC1) in the model alga
Chlamydomonas reinhardtii. We find that EPYC1 consists of five evenly spaced
Rubisco-binding regions that share sequence similarity. Single-particle
cryo-electron microscopy of these regions in complex with Rubisco indicates that
each Rubisco holoenzyme has eight binding sites for EPYC1, one on each Rubisco
small subunit. Interface mutations disrupt binding, phase separation and
pyrenoid formation. Cryo-electron tomography supports a model in which EPYC1 and
Rubisco form a codependent multivalent network of specific low-affinity bonds,
giving the matrix liquid-like properties. Our results advance the structural and
functional understanding of the phase separation underlying the pyrenoid, an
organelle that plays a fundamental role in the global carbon cycle.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|

