 |
PDBsum entry 7f1v
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of pseudomonas putida methionine gamma-lyase q349s mutant with l-homocysteine intermediates
|
|
Structure:
|
 |
L-methionine gamma-lyase. Chain: a, c. Synonym: mgl,homocysteine desulfhydrase,l-methioninase. Engineered: yes. Mutation: yes. L-methionine gamma-lyase. Chain: b, d. Synonym: mgl,homocysteine desulfhydrase,l-methioninase. Engineered: yes.
|
|
Source:
|
 |
Pseudomonas putida. Arthrobacter siderocapsulatus. Organism_taxid: 303. Gene: mdea. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.190
|
R-free:
|
0.227
|
|
|
Authors:
|
 |
A.Okawa,H.Handa,E.Yasuda,M.Murota,D.Kudo,T.Tamura,T.Shiba,K.Inagaki
|
|
Key ref:
|
 |
A.Okawa
et al.
(2022).
Characterization and application of l-Methionine gamm q349s mutant enzyme with an enhanced activity toward l-Homocysteine..
J biosci bioeng,
133,
213.
PubMed id:
|
 |
|
Date:
|
 |
|
09-Jun-21
|
Release date:
|
20-Apr-22
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P13254
(MEGL_PSEPU) -
L-methionine gamma-lyase from Pseudomonas putida
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
398 a.a.
396 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.4.4.1.11
- methionine gamma-lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-methionine + H2O = methanethiol + 2-oxobutanoate + NH4+
|
 |
 |
 |
 |
 |
L-methionine
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
H2O
|
=
|
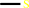 methanethiol
methanethiol
|
+
|
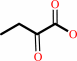 2-oxobutanoate
2-oxobutanoate
|
+
|
NH4(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
7XF)
matches with 62.50% similarity
|
|
 |
 |
Enzyme class 2:
|
 |
E.C.4.4.1.2
- homocysteine desulfhydrase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-homocysteine + H2O = 2-oxobutanoate + hydrogen sulfide + NH4+ + H+
|
 |
 |
 |
 |
 |
L-homocysteine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
=
|
 2-oxobutanoate
2-oxobutanoate
|
+
|
hydrogen sulfide
|
+
|
NH4(+)
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
7XF)
matches with 62.50% similarity
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

