 |
PDBsum entry 7ap2
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Neisseria gonorrhoeae leucyl-tRNA synthetase in complex with compound leus7hmdda
|
|
Structure:
|
 |
Leucine--tRNA ligase. Chain: a. Fragment: leucyl-tRNA synthetase. Synonym: leucyl-tRNA synthetase,leurs. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Neisseria gonorrhoeae. Organism_taxid: 485. Gene: leus, vt05_02036, whoo_00006, whoo_00455. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.25Å
|
R-factor:
|
0.190
|
R-free:
|
0.235
|
|
|
Authors:
|
 |
L.Pang,S.De Graef,S.V.Strelkov,S.D.Weeks
|
|
Key ref:
|
 |
B.Zhang
et al.
(2020).
Synthesis and biological evaluation of 1,3-Dideazapurine-Like 7-Amino-5-Hydroxymethyl-Benzim ribonucleoside analogues as aminoacyl-Trna synthetase inhibitors..
Molecules,
25,
.
PubMed id:
|
 |
|
Date:
|
 |
|
15-Oct-20
|
Release date:
|
28-Oct-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q5FAJ3
(SYL_NEIG1) -
Leucine--tRNA ligase from Neisseria gonorrhoeae (strain ATCC 700825 / FA 1090)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
876 a.a.
843 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.4
- leucine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Leu) + L-leucine + ATP = L-leucyl-tRNA(Leu) + AMP + diphosphate
|
 |
 |
 |
 |
 |
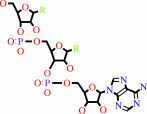 tRNA(Leu)
tRNA(Leu)
|
+
|
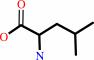 L-leucine
L-leucine
|
+
|
 ATP
ATP
|
=
|
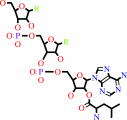 L-leucyl-tRNA(Leu)
L-leucyl-tRNA(Leu)
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

