 |
PDBsum entry 6v8c
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Design, synthesis, and mechanism of fluorine-substituted cyclohexene analogues of gama-aminobutyric acid (gaba) as selective ornithine aminotransferase inactivators
|
|
Structure:
|
 |
Ornithine aminotransferase, mitochondrial. Chain: a, b, c. Synonym: ornithine delta-aminotransferase,ornithine--oxo-acid aminotransferase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Gene: oat. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.240
|
R-free:
|
0.268
|
|
|
Authors:
|
 |
W.Zhu,P.T.Doubleday,D.S.Catlin,P.Weerawarna,A.Butrin,S.Shen, N.L.Kelleher,D.Liu,R.B.Silverman
|
|
Key ref:
|
 |
W.Zhu
et al.
Design, Synthesis, And mechanism of fluorine-Substitu cyclohexene analogues of gama-Aminobutyric acid (gaba selective ornithine aminotransferase inactivators.
To be published,
.
|
 |
|
Date:
|
 |
|
10-Dec-19
|
Release date:
|
16-Dec-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P04181
(OAT_HUMAN) -
Ornithine aminotransferase, mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
439 a.a.
404 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.6.1.13
- ornithine aminotransferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a 2-oxocarboxylate + L-ornithine = L-glutamate 5-semialdehyde + an L-alpha-amino acid
|
 |
 |
 |
 |
 |
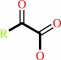 2-oxocarboxylate
2-oxocarboxylate
|
+
|
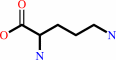 L-ornithine
L-ornithine
|
=
|
L-glutamate 5-semialdehyde
Bound ligand (Het Group name = )
matches with 58.33% similarity
|
+
|
L-alpha-amino acid
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
Pyridoxal 5'-phosphate
Bound ligand (Het Group name =
PLP)
matches with 93.75% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

