 |
PDBsum entry 6r2c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
6r2c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of the suca domain of mycobacterium smegmatis kgd after soaking with succinylphosphonate phosphonoethyl ester (pesp)
|
|
Structure:
|
 |
Multifunctional 2-oxoglutarate metabolism enzyme. Chain: a, b, c, d. Synonym: 2-hydroxy-3-oxoadipate synthase,hoas,2-oxoglutarate carboxy- lyase,2-oxoglutarate decarboxylase,alpha-ketoglutarate decarboxylase, kgd,alpha-ketoglutarate-glyoxylate carboligase. Ec: 2.2.1.5,4.1.1.71,1.2.4.2,2.3.1.61. Engineered: yes
|
|
Source:
|
 |
Mycobacterium smegmatis (strain atcc 700084 / mc(2)155). Organism_taxid: 246196. Strain: atcc 700084 / mc(2)155. Gene: kgd, suca, msmeg_5049, msmei_4922. Expressed in: escherichia coli bl21(de3). Expression_system_taxid: 469008
|
|
Resolution:
|
 |
|
2.09Å
|
R-factor:
|
0.233
|
R-free:
|
0.246
|
|
|
Authors:
|
 |
T.Wagner,P.M.Alzari,M.Bellinzoni
|
|
Key ref:
|
 |
T.Wagner
et al.
(2019).
Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
J Struct Biol,
208,
182-190.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
15-Mar-19
|
Release date:
|
11-Sep-19
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
A0R2B1
(KGD_MYCS2) -
Multifunctional 2-oxoglutarate metabolism enzyme from Mycolicibacterium smegmatis (strain ATCC 700084 / mc(2)155)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1227 a.a.
814 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.2.4.2
- oxoglutarate dehydrogenase (succinyl-transferring).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Oxo-acid dehydrogenase complexes
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-[(R)-lipoyl]-L-lysyl-[protein] + 2-oxoglutarate + H+ = N6-[(R)- S(8)-succinyldihydrolipoyl]-L-lysyl-[protein] + CO2
|
 |
 |
 |
 |
 |
N(6)-[(R)-lipoyl]-L-lysyl-[protein]
|
+
|
2-oxoglutarate
Bound ligand (Het Group name = )
matches with 43.75% similarity
|
+
|
H(+)
|
=
|
N(6)-[(R)- S(8)-succinyldihydrolipoyl]-L-lysyl-[protein]
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
TPP)
corresponds exactly
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.2.2.1.5
- 2-hydroxy-3-oxoadipate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
glyoxylate + 2-oxoglutarate + H+ = 2-hydroxy-3-oxoadipate + CO2
|
 |
 |
 |
 |
 |
glyoxylate
Bound ligand (Het Group name = )
matches with 43.75% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
H(+)
|
=
|
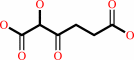 2-hydroxy-3-oxoadipate
2-hydroxy-3-oxoadipate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
TPP)
corresponds exactly
|
|
 |
 |
Enzyme class 4:
|
 |
E.C.2.3.1.61
- dihydrolipoyllysine-residue succinyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-[(R)-dihydrolipoyl]-L-lysyl-[protein] + succinyl-CoA = N6-[(R)- S(8)-succinyldihydrolipoyl]-L-lysyl-[protein] + CoA
|
 |
 |
 |
 |
 |
N(6)-[(R)-dihydrolipoyl]-L-lysyl-[protein]
|
+
|
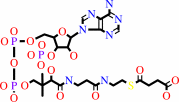 succinyl-CoA
succinyl-CoA
|
=
|
N(6)-[(R)- S(8)-succinyldihydrolipoyl]-L-lysyl-[protein]
|
+
|
 CoA
CoA
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 5:
|
 |
E.C.4.1.1.71
- 2-oxoglutarate decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-oxoglutarate + H+ = succinate semialdehyde + CO2
|
 |
 |
 |
 |
 |
 2-oxoglutarate
2-oxoglutarate
|
+
|
H(+)
|
=
|
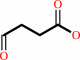 succinate semialdehyde
succinate semialdehyde
|
+
|
CO2
Bound ligand (Het Group name = )
matches with 53.85% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Thiamine diphosphate
|
 |
 |
 |
 |
 |
Thiamine diphosphate
Bound ligand (Het Group name =
TPP)
corresponds exactly
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Struct Biol
208:182-190
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
Conformational transitions in the active site of mycobacterial 2-oxoglutarate dehydrogenase upon binding phosphonate analogues of 2-oxoglutarate: From a Michaelis-like complex to ThDP adducts.
|
|
T.Wagner,
A.Boyko,
P.M.Alzari,
V.I.Bunik,
M.Bellinzoni.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Mycobacterial KGD, the thiamine diphosphate (ThDP)-dependent E1o component of
the 2-oxoglutarate dehydrogenase complex (OGDHC), is known to undergo
significant conformational changes during catalysis with two distinct
conformational states, previously named as the early and late state. In this
work, we employ two phosphonate analogues of 2-oxoglutarate (OG), i.e. succinyl
phosphonate (SP) and phosphono ethyl succinyl phosphonate (PESP), as tools to
isolate the first catalytic steps and understand the significance of
conformational transitions for the enzyme regulation. The kinetics showed a more
efficient inhibition of mycobacterial E1o by SP (Ki
0.043 ± 0.013 mM) than PESP (Ki 0.88 ± 0.28 mM),
consistent with the different circular dichroism spectra of the corresponding
complexes. PESP allowed us to get crystallographic snapshots of the
Michaelis-like complex, the first one for 2-oxo acid dehydrogenases, followed by
the covalent adduction of the inhibitor to ThDP, mimicking the
pre-decarboxylation complex. In addition, covalent ThDP-phosphonate complexes
obtained with both compounds by co-crystallization were in the late
conformational state, probably corresponding to slowly dissociating
enzyme-inhibitor complexes. We discuss the relevance of these findings in terms
of regulatory features of the mycobacterial E1o enzymes, and in the perspective
of developing tools for species-specific metabolic regulation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

