 |
PDBsum entry 6lyh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of tea n9-methyltransferase cktcs in complex with sah and 1,3,7-trimethyluric acid
|
|
Structure:
|
 |
N-methyltransferase cktcs. Chain: b, a, c, d, e, f, g, h. Engineered: yes
|
|
Source:
|
 |
Camellia sinensis var. Assamica. Organism_taxid: 261999. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
3.15Å
|
R-factor:
|
0.264
|
R-free:
|
0.298
|
|
|
Authors:
|
 |
Y.Wang,Z.-M.Zhang
|
|
Key ref:
|
 |
Y.H.Zhang
et al.
(2020).
Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea.
Nat Commun,
11,
1473.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
14-Feb-20
|
Release date:
|
04-Mar-20
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9FZN8
(TCS1_CAMSI) -
3,7-dimethylxanthine N-methyltransferase TCS1 from Camellia sinensis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
369 a.a.
350 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|
*
PDB and UniProt seqs differ
at 27 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.1.160
- caffeine synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
theobromine + S-adenosyl-L-methionine = caffeine + S-adenosyl-L- homocysteine + H+
|
|
2.
|
1,7-dimethylxanthine + S-adenosyl-L-methionine = caffeine + S-adenosyl-L-homocysteine + H+
|
|
3.
|
7-methylxanthine + S-adenosyl-L-methionine = theobromine + S-adenosyl-L-homocysteine + H+
|
|
 |
 |
 |
 |
 |
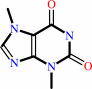 theobromine
theobromine
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
caffeine
Bound ligand (Het Group name = )
matches with 93.33% similarity
|
+
|
 S-adenosyl-L- homocysteine
S-adenosyl-L- homocysteine
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
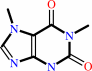 1,7-dimethylxanthine
1,7-dimethylxanthine
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
caffeine
Bound ligand (Het Group name = )
matches with 93.33% similarity
|
+
|
 S-adenosyl-L-homocysteine
S-adenosyl-L-homocysteine
|
+
|
H(+)
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
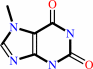 7-methylxanthine
7-methylxanthine
|
+
|
 S-adenosyl-L-methionine
S-adenosyl-L-methionine
|
=
|
 theobromine
theobromine
|
+
|
S-adenosyl-L-homocysteine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 86.67% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Commun
11:1473
(2020)
|
|
PubMed id:
|
|
|
|

|
| |
|
Identification and characterization of N9-methyltransferase involved in converting caffeine into non-stimulatory theacrine in tea.
|
|
Y.H.Zhang,
Y.F.Li,
Y.Wang,
L.Tan,
Z.Q.Cao,
C.Xie,
G.Xie,
H.B.Gong,
W.Y.Sun,
S.H.Ouyang,
W.J.Duan,
X.Lu,
K.Ding,
H.Kurihara,
D.Hu,
Z.M.Zhang,
I.Abe,
R.R.He.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Caffeine is a major component of xanthine alkaloids and commonly consumed in
many popular beverages. Due to its occasional side effects, reduction of
caffeine in a natural way is of great importance and economic significance.
Recent studies reveal that caffeine can be converted into non-stimulatory
theacrine in the rare tea plant Camellia assamica var. kucha (Kucha), which
involves oxidation at the C8 and methylation at the N9 positions of caffeine.
However, the underlying molecular mechanism remains unclear. Here, we identify
the theacrine synthase CkTcS from Kucha, which possesses novel
N9-methyltransferase activity using 1,3,7-trimethyluric acid but not caffeine as
a substrate, confirming that C8 oxidation takes place prior to N9-methylation.
The crystal structure of the CkTcS complex reveals the key residues that are
required for the N9-methylation, providing insights into how caffeine
N-methyltransferases in tea plants have evolved to catalyze regioselective
N-methylation through fine tuning of their active sites. These results may guide
the future development of decaffeinated drinks.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

