 |
PDBsum entry 6lpy

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
6lpy
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.3.8.1
- short-chain acyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a short-chain 2,3-saturated fatty acyl-CoA + oxidized [electron-transfer flavoprotein] + H+ = a short-chain (2E)-enoyl-CoA + reduced [electron- transfer flavoprotein]
|
 |
 |
 |
 |
 |
Butanoyl-CoA
Bound ligand (Het Group name = )
matches with 90.57% similarity
|
+
|
electron-transfer flavoprotein
|
=
|
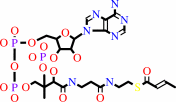 2-butenoyl-CoA
2-butenoyl-CoA
|
+
|
reduced electron-transfer flavoprotein
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
Enzyme class 3:
|
 |
E.C.1.3.8.7
- medium-chain acyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a medium-chain 2,3-saturated fatty acyl-CoA + oxidized [electron-transfer flavoprotein] + H+ = a medium-chain (2E)-enoyl-CoA + reduced [electron- transfer flavoprotein]
|
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
E.C.1.3.8.8
- long-chain-acyl-CoA dehydrogenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a long-chain 2,3-saturated fatty acyl-CoA + oxidized [electron-transfer flavoprotein] + H+ = a long-chain (2E)-enoyl-CoA + reduced [electron- transfer flavoprotein]
|
 |
 |
 |
 |
 |
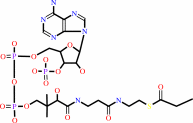 Long-chain-acyl-CoA
Long-chain-acyl-CoA
|
+
|
electron-transfer flavoprotein
|
=
|
long-chain-2,3- dehydroacyl-CoA
|
+
|
reduced electron-transfer flavoprotein
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
117:16324-16332
(2020)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural basis for the broad substrate specificity of two acyl-CoA dehydrogenases FadE5 from mycobacteria.
|
|
X.Chen,
J.Chen,
B.Yan,
W.Zhang,
L.W.Guddat,
X.Liu,
Z.Rao.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
FadE, an acyl-CoA dehydrogenase, introduces unsaturation to carbon chains in
lipid metabolism pathways. Here, we report that FadE5 from Mycobacterium
tuberculosis (MtbFadE5) and Mycobacterium smegmatis
(MsFadE5) play roles in drug resistance and exhibit broad specificity for
linear acyl-CoA substrates but have a preference for those with long carbon
chains. Here, the structures of MsFadE5 and MtbFadE5, in the
presence and absence of substrates, have been determined. These reveal the
molecular basis for the broad substrate specificity of these enzymes. FadE5
interacts with the CoA region of the substrate through a large number of
hydrogen bonds and an unusual π-π stacking interaction, allowing these enzymes
to accept both short- and long-chain substrates. Residues in the substrate
binding cavity reorient their side chains to accommodate substrates of various
lengths. Longer carbon-chain substrates make more numerous hydrophobic
interactions with the enzyme compared with the shorter-chain substrates,
resulting in a preference for this type of substrate.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

