 |
PDBsum entry 6hv6
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.2.4.1.-
- ?????
|
|
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.3.5.1.44
- protein-glutamine glutaminase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-glutaminyl-[protein] + H2O = L-glutamyl-[protein] + NH4+
|
 |
 |
 |
 |
 |
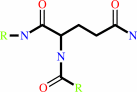 Protein L-glutamine
Protein L-glutamine
|
+
|
H(2)O
|
=
|
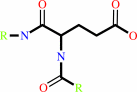 protein L-glutamate
protein L-glutamate
|
+
|
NH(3)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
294:1035-1044
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
A cysteine protease-like domain enhances the cytotoxic effects of the Photorhabdus asymbiotica toxin PaTox.
|
|
X.Bogdanovic,
S.Schneider,
N.Levanova,
C.Wirth,
C.Trillhaase,
M.Steinemann,
C.Hunte,
K.Aktories,
T.Jank.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The nematode mutualistic bacterium Photorhabdus asymbiotica produces a
large virulence-associated multifunctional protein toxin named PaTox. A
glycosyltransferase domain and a deamidase domain of this large toxin function
as effectors that specifically target host Rho GTPases and heterotrimeric G
proteins, respectively. Modification of these intracellular regulators results
in toxicity toward insects and mammalian cells. In this study, we identified a
cysteine protease-like domain spanning PaTox residues 1844-2114
(PaToxP), upstream of these two effector domains and characterized by
three conserved amino acid residues (Cys-1865, His-1955, and Asp-1975). We
determined the crystal structure of the PaToxP C1865A variant by
native single-wavelength anomalous diffraction of sulfur atoms (sulfur-SAD). At
2.0 Å resolution, this structure revealed a catalytic site typical for
papain-like cysteine proteases, comprising a catalytic triad, oxyanion hole, and
typical secondary structural elements. The PaToxP structure had
highest similarity to that of the AvrPphB protease from Pseudomonas
syringae classified as a C58-protease. Furthermore, we observed that
PaToxP shares structural homology also with non-C58-cysteine
proteases, deubiquitinases, and deamidases. Upon delivery into insect larvae,
PaToxP alone without full-length PaTox had no toxic effects. Yet,
PaToxP expression in mammalian cells was toxic and enhanced the
apoptotic phenotype induced by PaTox in HeLa cells. We propose that
PaToxP is a C58-like cysteine protease module that is essential for
full PaTox activity.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

