 |
PDBsum entry 6hu9

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase/electron transport
|
PDB id
|
|

|

|
6hu9
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|

 431 a.a.
431 a.a.
|
 |

|

|

|

|

|

|

|

 352 a.a.
352 a.a.
|
 |

|

|

|

|

|

|

|

 385 a.a.
385 a.a.
|
 |

|

|

|

|

|

|

|

 247 a.a.
247 a.a.
|
 |

|

|

|

|

|

|

|

 185 a.a.
185 a.a.
|
 |

|

|

|

|

|

|

|

 75 a.a.
75 a.a.
|
 |

|

|

|

|

|

|

|

 126 a.a.
126 a.a.
|
 |

|

|

|

|

|

|

|

 93 a.a.
93 a.a.
|
 |

|

|

|

|

|

|

|

 57 a.a.
57 a.a.
|
 |

|

|

|

|

|

|

|

 76 a.a.
76 a.a.
|
 |

|

|

|

|

|

|

|

 534 a.a.
534 a.a.
|
 |

|

|

|

|

|

|

|

 236 a.a.
236 a.a.
|
 |

|

|

|

|

|

|

|

 269 a.a.
269 a.a.
|
 |

|

|

|

|

|

|

|

 121 a.a.
121 a.a.
|
 |

|

|

|

|

|

|

|

 133 a.a.
133 a.a.
|
 |

|

|

|

|

|

|

|

 102 a.a.
102 a.a.
|
 |

|

|

|

|

|

|

|

 59 a.a.
59 a.a.
|
 |

|

|

|

|

|

|

|

 47 a.a.
47 a.a.
|
 |

|

|

|

|

|

|

|

 55 a.a.
55 a.a.
|
 |

|

|

|

|

|

|

|

 77 a.a.
77 a.a.
|
 |

|

|

|

|

|

|

|

 113 a.a.
113 a.a.
|
 |

|

|

|

|

|

|

|

 45 a.a.
45 a.a.
|
 |

|

|
|
|
|

|

|

|

|

|
 _ZN
×2 _ZN
×2
|
 |

|

|

|

|

|

|

|
 _CU
×2 _CU
×2
|
 |

|

|

|

|

|

|

|
 _CA
×2 _CA
×2
|
 |

|

|

|

|

|

|

|
 _MG
×2 _MG
×2
|
 |

|

|

|

|

|

|

|
 CUA
×4 CUA
×4
|
 |

|

|
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase/electron transport
|
 |
|
Title:
|
 |
Iii2-iv2 mitochondrial respiratory supercomplex from s. Cerevisiae
|
|
Structure:
|
 |
Cytochrome b-c1 complex subunit 1, mitochondrial. Chain: a, l. Synonym: complex iii subunit 1,core protein i,ubiquinol-cytochromE-C reductase complex core protein 1. Cytochrome b-c1 complex subunit 2, mitochondrial. Chain: b, m. Synonym: complex iii subunit 2,core protein ii,ubiquinol-cytochromE-C reductase complex core protein 2. Cytochrome b.
|
|
Source:
|
 |
Saccharomyces cerevisiae (strain atcc 204508 / s288c). Baker's yeast. Organism_taxid: 559292. Strain: atcc 204508 / s288c. Gene: cox13, ygl191w, g1341. Expressed in: saccharomyces cerevisiae. Expression_system_taxid: 4932. Strain: atcc 204508 / s288c
|
|
Authors:
|
 |
A.M.Hartley,N.Pinotsis,A.Marechal
|
|
Key ref:
|
 |
A.M.Hartley
et al.
(2019).
Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1.
Nat Struct Mol Biol,
26,
78-83.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
05-Oct-18
|
Release date:
|
26-Dec-18
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P07256
(QCR1_YEAST) -
Cytochrome b-c1 complex subunit 1, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
457 a.a.
431 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07257
(QCR2_YEAST) -
Cytochrome b-c1 complex subunit 2, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
368 a.a.
352 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00163
(CYB_YEAST) -
Cytochrome b from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
385 a.a.
385 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07143
(CY1_YEAST) -
Cytochrome c1, heme protein, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
309 a.a.
247 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P08067
(UCRI_YEAST) -
Cytochrome b-c1 complex subunit Rieske, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
215 a.a.
185 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00127
(QCR6_YEAST) -
Cytochrome b-c1 complex subunit 6, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
147 a.a.
75 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00128
(QCR7_YEAST) -
Cytochrome b-c1 complex subunit 7, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
127 a.a.
126 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P08525
(QCR8_YEAST) -
Cytochrome b-c1 complex subunit 8, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
94 a.a.
93 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P22289
(QCR9_YEAST) -
Cytochrome b-c1 complex subunit 9, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
66 a.a.
57 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P37299
(QCR10_YEAST) -
Cytochrome b-c1 complex subunit 10, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
77 a.a.
76 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00401
(COX1_YEAST) -
Cytochrome c oxidase subunit 1 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
534 a.a.
534 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00410
(COX2_YEAST) -
Cytochrome c oxidase subunit 2 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
251 a.a.
236 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00420
(COX3_YEAST) -
Cytochrome c oxidase subunit 3 from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
269 a.a.
269 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P04037
(COX4_YEAST) -
Cytochrome c oxidase subunit 4, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
155 a.a.
121 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00424
(COX5A_YEAST) -
Cytochrome c oxidase subunit 5A, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
153 a.a.
133 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P00427
(COX6_YEAST) -
Cytochrome c oxidase subunit 6, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
148 a.a.
102 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P10174
(COX7_YEAST) -
Cytochrome c oxidase subunit 7, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
60 a.a.
59 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P04039
(COX8_YEAST) -
Cytochrome c oxidase subunit 8, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
78 a.a.
47 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
P07255
(COX9_YEAST) -
Cytochrome c oxidase subunit 9, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
59 a.a.
55 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|

|

|
|
Q01519
(COX12_YEAST) -
Cytochrome c oxidase subunit 12, mitochondrial from Saccharomyces cerevisiae (strain ATCC 204508 / S288c)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
83 a.a.
77 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
Chains a, b, c, m, n, o:
E.C.7.1.1.9
- cytochrome-c oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4 Fe(II)-[cytochrome c] + O2 + 8 H+(in) = 4 Fe(III)-[cytochrome c] + 2 H2O + 4 H+(out)
|
 |
 |
 |
 |
 |
4
×
Fe(II)-[cytochrome c]
|
+
|
 O2
O2
|
+
|
8
×
H(+)(in)
|
=
|
4
×
Fe(III)-[cytochrome c]
|
+
|
2
×
H2O
|
+
|
4
×
H(+)(out)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
Chains C, D, E, N, O, P:
E.C.7.1.1.8
- quinol--cytochrome-c reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a quinol + 2 Fe(III)-[cytochrome c](out) = a quinone + 2 Fe(II)- [cytochrome c](out) + 2 H(+)(out)
|
 |
 |
 |
 |
 |
4
×
quinol
|
+
|
2
×
Fe(III)-[cytochrome c](out)
|
=
|
 8
×
quinone
8
×
quinone
|
+
|
2
×
Fe(II)- [cytochrome c](out)
|
+
|
2
×
H(+)(out)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 4:
|
 |
Chains d, e, f, g, h, i, j, k, p, q, r, s, t, u, v, w:
E.C.1.9.3.1
- Transferred entry: 7.1.1.9.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4 ferrocytochrome c + O2 + 4 H+ = 4 ferricytochrome c + 2 H2O
|
 |
 |
 |
 |
 |
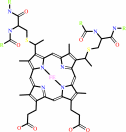 4
×
ferrocytochrome c
4
×
ferrocytochrome c
|
+
|
 2
×
O(2)
2
×
O(2)
|
+
|
4
×
H(+)
|
=
|
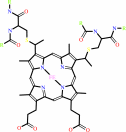 4
×
ferricytochrome c
4
×
ferricytochrome c
|
+
|
2
×
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nat Struct Mol Biol
26:78-83
(2019)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of yeast cytochrome c oxidase in a supercomplex with cytochrome bc1.
|
|
A.M.Hartley,
N.Lukoyanova,
Y.Zhang,
A.Cabrera-Orefice,
S.Arnold,
B.Meunier,
N.Pinotsis,
A.Maréchal.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Cytochrome c oxidase (complex IV, CIV) is known in mammals to exist
independently or in association with other respiratory proteins to form
supercomplexes (SCs). In Saccharomyces cerevisiae, CIV is found solely in an SC
with cytochrome bc1 (complex III, CIII). Here, we present the
cryogenic electron microscopy (cryo-EM) structure of S. cerevisiae CIV in a
III2IV2 SC at 3.3 Å resolution. While overall
similarity to mammalian homologs is high, we found notable differences in the
supernumerary subunits Cox26 and Cox13; the latter exhibits a unique arrangement
that precludes CIV dimerization as seen in bovine. A conformational shift in the
matrix domain of Cox5A-involved in allosteric inhibition by ATP-may arise from
its association with CIII. The CIII-CIV arrangement highlights a conserved
interaction interface of CIII, albeit one occupied by complex I in mammalian
respirasomes. We discuss our findings in the context of the potential impact of
SC formation on CIV regulation.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |

| | |

