 |
PDBsum entry 6amc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Biosynthetic protein
|
PDB id
|
|

|

|
6amc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.20
- tryptophan synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate + L-serine = D-glyceraldehyde 3-phosphate + L-tryptophan + H2O
|
 |
 |
 |
 |
 |
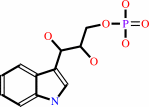 (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
 L-serine
L-serine
|
=
|
 D-glyceraldehyde 3-phosphate
D-glyceraldehyde 3-phosphate
|
+
|
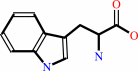 L-tryptophan
L-tryptophan
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Am Chem Soc
140:7256-7266
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Directed Evolution Mimics Allosteric Activation by Stepwise Tuning of the Conformational Ensemble.
|
|
A.R.Buller,
P.van Roye,
J.K.B.Cahn,
R.A.Scheele,
M.Herger,
F.H.Arnold.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Allosteric enzymes contain a wealth of catalytic diversity that remains
distinctly underutilized for biocatalysis. Tryptophan synthase is a model
allosteric system and a valuable enzyme for the synthesis of noncanonical amino
acids (ncAA). Previously, we evolved the β-subunit from Pyrococcus furiosus,
PfTrpB, for ncAA synthase activity in the absence of its native partner protein
PfTrpA. However, the precise mechanism by which mutation activated TrpB to
afford a stand-alone catalyst remained enigmatic. Here, we show that directed
evolution caused a gradual change in the rate-limiting step of the catalytic
cycle. Concomitantly, the steady-state distribution of the intermediates shifts
to favor covalently bound Trp adducts, which have increased thermodynamic
stability. The biochemical properties of these evolved, stand-alone TrpBs
converge on those induced in the native system by allosteric activation.
High-resolution crystal structures of the wild-type enzyme, an intermediate in
the lineage, and the final variant, encompassing five distinct chemical states,
show that activating mutations have only minor structural effects on their
immediate environment. Instead, mutation stabilizes the large-scale motion of a
subdomain to favor an otherwise transiently populated closed conformational
state. This increase in stability enabled the first structural description of
Trp covalently bound in a catalytically active TrpB, confirming key features of
catalysis. These data combine to show that sophisticated models of allostery are
not a prerequisite to recapitulating its complex effects via directed evolution,
opening the way to engineering stand-alone versions of diverse allosteric
enzymes.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

