 |
PDBsum entry 6evm
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.11.2
- procollagen-proline 4-dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
L-prolyl-[collagen] + 2-oxoglutarate + O2 = trans-4-hydroxy-L-prolyl- [collagen] + succinate + CO2
|
 |
 |
 |
 |
 |
L-prolyl-[collagen]
Bound ligand (Het Group name = )
matches with 63.64% similarity
|
+
|
 2-oxoglutarate
2-oxoglutarate
|
+
|
 O2
O2
|
=
|
trans-4-hydroxy-L-prolyl- [collagen]
|
+
|
 succinate
succinate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Fe(2+); L-ascorbate
|
 |
 |
 |
 |
 |
Fe(2+)
|
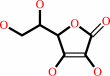 L-ascorbate
L-ascorbate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Protein Sci
27:1692-1703
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural enzymology binding studies of the peptide-substrate-binding domain of human collagen prolyl 4-hydroxylase (type-II): High affinity peptides have a PxGP sequence motif.
|
|
A.V.Murthy,
R.Sulu,
M.K.Koski,
H.Tu,
J.Anantharajan,
S.K.Sah-Teli,
J.Myllyharju,
R.K.Wierenga.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The peptide-substrate-binding (PSB) domain of collagen prolyl 4-hydroxylase
(C-P4H, an α2 β2 tetramer) binds proline-rich
procollagen peptides. This helical domain (the middle domain of the α subunit)
has an important role concerning the substrate binding properties of C-P4H,
although it is not known how the PSB domain influences the hydroxylation
properties of the catalytic domain (the C-terminal domain of the α subunit).
The crystal structures of the PSB domain of the human C-P4H isoform II (PSB-II)
complexed with and without various short proline-rich peptides are described.
The comparison with the previously determined PSB-I peptide complex structures
shows that the C-P4H-I substrate peptide (PPG)3 , has at most very
weak affinity for PSB-II, although it binds with high affinity to PSB-I. The
replacement of the middle PPG triplet of (PPG)3 to the
nonhydroxylatable PAG, PRG, or PEG triplet, increases greatly the affinity of
PSB-II for these peptides, leading to a deeper mode of binding, as compared to
the previously determined PSB-I peptide complexes. In these PSB-II complexes,
the two peptidyl prolines of its central P(A/R/E)GP region bind in the Pro5 and
Pro8 binding pockets of the PSB peptide-binding groove, and direct hydrogen
bonds are formed between the peptide and the side chains of the highly conserved
residues Tyr158, Arg223, and Asn227, replacing water mediated interactions in
the corresponding PSB-I complex. These results suggest that PxGP (where x is not
a proline) is the common motif of proline-rich peptide sequences that bind with
high affinity to PSB-II.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

