 |
PDBsum entry 5dw3
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.20
- tryptophan synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Tryptophan Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate + L-serine = D-glyceraldehyde 3-phosphate + L-tryptophan + H2O
|
 |
 |
 |
 |
 |
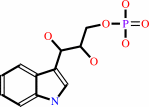 (1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
(1S,2R)-1-C-(indol-3-yl)glycerol 3-phosphate
|
+
|
 L-serine
L-serine
|
=
|
D-glyceraldehyde 3-phosphate
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
L-tryptophan
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Pyridoxal 5'-phosphate
|
 |
 |
 |
 |
 |
 Pyridoxal 5'-phosphate
Pyridoxal 5'-phosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
112:14599-14604
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Directed evolution of the tryptophan synthase β-subunit for stand-alone function recapitulates allosteric activation.
|
|
A.R.Buller,
S.Brinkmann-Chen,
D.K.Romney,
M.Herger,
J.Murciano-Calles,
F.H.Arnold.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Enzymes in heteromeric, allosterically regulated complexes catalyze a rich array
of chemical reactions. Separating the subunits of such complexes, however, often
severely attenuates their catalytic activities, because they can no longer be
activated by their protein partners. We used directed evolution to explore
allosteric regulation as a source of latent catalytic potential using the
β-subunit of tryptophan synthase from Pyrococcus furiosus (PfTrpB). As part of
its native αββα complex, TrpB efficiently produces tryptophan and tryptophan
analogs; activity drops considerably when it is used as a stand-alone catalyst
without the α-subunit. Kinetic, spectroscopic, and X-ray crystallographic data
show that this lost activity can be recovered by mutations that reproduce the
effects of complexation with the α-subunit. The engineered PfTrpB is a powerful
platform for production of Trp analogs and for further directed evolution to
expand substrate and reaction scope.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

