 |
PDBsum entry 5dot
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of human carbamoyl phosphate synthetase i (cps1), apo form
|
|
Structure:
|
 |
Carbamoyl-phosphate synthase [ammonia], mitochondrial. Chain: a, b. Fragment: mature enzyme. Synonym: carbamoyl-phosphate synthetase i,cpsase i. Engineered: yes. Other_details: mature cps1 with n-terminal his-tag for purification purposes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Organ: liver. Gene: cps1. Expressed in: spodoptera frugiperda. Expression_system_taxid: 7108. Expression_system_cell_line: sf9.
|
|
Resolution:
|
 |
|
2.40Å
|
R-factor:
|
0.166
|
R-free:
|
0.196
|
|
|
Authors:
|
 |
L.M.Polo,S.De Cima,I.Fita,V.Rubio
|
|
Key ref:
|
 |
S.de Cima
et al.
(2015).
Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
Sci Rep,
5,
16950.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
11-Sep-15
|
Release date:
|
09-Dec-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P31327
(CPSM_HUMAN) -
Carbamoyl-phosphate synthase [ammonia], mitochondrial from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
1500 a.a.
1352 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.3.4.16
- carbamoyl-phosphate synthase (ammonia).
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogencarbonate + NH4+ + 2 ATP = carbamoyl phosphate + 2 ADP + phosphate + 2 H+
|
 |
 |
 |
 |
 |
 hydrogencarbonate
hydrogencarbonate
|
+
|
NH4(+)
|
+
|
 2
×
ATP
2
×
ATP
|
=
|
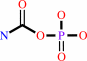 carbamoyl phosphate
carbamoyl phosphate
|
+
|
 2
×
ADP
2
×
ADP
|
+
|
 phosphate
phosphate
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Sci Rep
5:16950
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of human carbamoyl phosphate synthetase: deciphering the on/off switch of human ureagenesis.
|
|
S.de Cima,
L.M.Polo,
C.Díez-Fernández,
A.I.Martínez,
J.Cervera,
I.Fita,
V.Rubio.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human carbamoyl phosphate synthetase (CPS1), a 1500-residue multidomain enzyme,
catalyzes the first step of ammonia detoxification to urea requiring
N-acetyl-L-glutamate (NAG) as essential activator to prevent ammonia/amino acids
depletion. Here we present the crystal structures of CPS1 in the absence and in
the presence of NAG, clarifying the on/off-switching of the urea cycle by NAG.
By binding at the C-terminal domain of CPS1, NAG triggers long-range
conformational changes affecting the two distant phosphorylation domains. These
changes, concerted with the binding of nucleotides, result in a dramatic
remodeling that stabilizes the catalytically competent conformation and the
building of the ~35 Å-long tunnel that allows migration of the carbamate
intermediate from its site of formation to the second phosphorylation site,
where carbamoyl phosphate is produced. These structures allow rationalizing the
effects of mutations found in patients with CPS1 deficiency (presenting
hyperammonemia, mental retardation and even death), as exemplified here for some
mutations.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

