 |
PDBsum entry 4zqb

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4zqb
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of NADP-dependent dehydrogenase from rhodobactersphaeroides in complex with NADP and sulfate
|
|
Structure:
|
 |
NADP-dependent dehydrogenase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Rhodobacter sphaeroides (strain atcc 17023 / 2.4.1 / ncib 8253 / dsm 158). Organism_taxid: 272943. Strain: atcc 17023 / 2.4.1 / ncib 8253 / dsm 158. Gene: rsp_3442. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.85Å
|
R-factor:
|
0.167
|
R-free:
|
0.200
|
|
|
Authors:
|
 |
M.Kowiel,O.A.Gasiorowska,I.G.Shabalin,K.B.Handing,P.J.Porebski, J.Bonanno,S.C.Almo,W.Minor,New York Structural Genomics Research Consortium (Nysgrc)
|
|
Key ref:
|
 |
M.Kowiel
et al.
Crystal structure of NADP-Dependent dehydrogenase fro rhodobactersphaeroides in complex with NADP and sulfa.
To be published,
.
|
 |
|
Date:
|
 |
|
08-May-15
|
Release date:
|
20-May-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q3IWN8
(Q3IWN8_RHOS4) -
2-hydroxyacid dehydrogenase from Cereibacter sphaeroides (strain ATCC 17023 / DSM 158 / JCM 6121 / CCUG 31486 / LMG 2827 / NBRC 12203 / NCIMB 8253 / ATH 2.4.1.)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
313 a.a.
313 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.1.1.1.79
- glyoxylate reductase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
glycolate + NADP+ = glyoxylate + NADPH + H+
|
 |
 |
 |
 |
 |
glycolate
Bound ligand (Het Group name = )
matches with 57.14% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
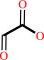 glyoxylate
glyoxylate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.1.1.1.81
- hydroxypyruvate reductase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
(R)-glycerate + NAD+ = 3-hydroxypyruvate + NADH + H+
|
|
2.
|
(R)-glycerate + NADP+ = 3-hydroxypyruvate + NADPH + H+
|
|
 |
 |
 |
 |
 |
(R)-glycerate
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
+
|
NAD(+)
Bound ligand (Het Group name = )
matches with 91.67% similarity
|
=
|
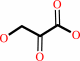 3-hydroxypyruvate
3-hydroxypyruvate
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
(R)-glycerate
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
 3-hydroxypyruvate
3-hydroxypyruvate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

