 |
PDBsum entry 4z0p

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4z0p
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of NADPH-dependent glyoxylate/hydroxypyruvate reductase smc02828 (smghra) from sinorhizobium meliloti in complex with NADPH and oxalate
|
|
Structure:
|
 |
NAD-dependent dehydrogenase. Chain: a. Engineered: yes
|
|
Source:
|
 |
Rhizobium meliloti (strain 1021). Ensifer meliloti. Organism_taxid: 266834. Strain: 1021. Gene: r00152, smc02828. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.70Å
|
R-factor:
|
0.135
|
R-free:
|
0.150
|
|
|
Authors:
|
 |
P.Sroka,O.A.Gasiorowska,K.B.Handing,I.G.Shabalin,P.J.Porebski, B.S.Hillerich,J.Bonanno,S.C.Almo,W.Minor,New York Structural Genomics Research Consortium (Nysgrc)
|
|
Key ref:
|
 |
J.Kutner
et al.
(2018).
Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
Biochemistry,
57,
963-977.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
26-Mar-15
|
Release date:
|
08-Apr-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q92T34
(Q92T34_RHIME) -
NAD-dependent dehydrogenase from Rhizobium meliloti (strain 1021)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
319 a.a.
316 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.79
- glyoxylate reductase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
glycolate + NADP+ = glyoxylate + NADPH + H+
|
 |
 |
 |
 |
 |
glycolate
Bound ligand (Het Group name = )
matches with 83.33% similarity
|
+
|
NADP(+)
Bound ligand (Het Group name = )
corresponds exactly
|
=
|
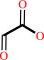 glyoxylate
glyoxylate
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
57:963-977
(2018)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural, Biochemical, and Evolutionary Characterizations of Glyoxylate/Hydroxypyruvate Reductases Show Their Division into Two Distinct Subfamilies.
|
|
J.Kutner,
I.G.Shabalin,
D.Matelska,
K.B.Handing,
O.Gasiorowska,
P.Sroka,
M.W.Gorna,
K.Ginalski,
K.Wozniak,
W.Minor.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The d-2-hydroxyacid dehydrogenase (2HADH) family illustrates a complex
evolutionary history with multiple lateral gene transfers and gene duplications
and losses. As a result, the exact functional annotation of individual members
can be extrapolated to a very limited extent. Here, we revise the previous
simplified view on the classification of the 2HADH family; specifically, we show
that the previously delineated glyoxylate/hydroxypyruvate reductase (GHPR)
subfamily consists of two evolutionary separated GHRA and GHRB subfamilies. We
compare two representatives of these subfamilies from Sinorhizobium meliloti
(SmGhrA and SmGhrB), employing a combination of biochemical, structural, and
bioinformatics approaches. Our kinetic results show that both enzymes reduce
several 2-ketocarboxylic acids with overlapping, but not equivalent, substrate
preferences. SmGhrA and SmGhrB show highest activity with glyoxylate and
hydroxypyruvate, respectively; in addition, only SmGhrB reduces
2-keto-d-gluconate, and only SmGhrA reduces pyruvate (with low efficiency). We
present nine crystal structures of both enzymes in apo forms and in complexes
with cofactors and substrates/substrate analogues. In particular, we determined
a crystal structure of SmGhrB with 2-keto-d-gluconate, which is the biggest
substrate cocrystallized with a 2HADH member. The structures reveal significant
differences between SmGhrA and SmGhrB, both in the overall structure and within
the substrate-binding pocket, offering insight into the molecular basis for the
observed substrate preferences and subfamily differences. In addition, we
provide an overview of all GHRA and GHRB structures complexed with a ligand in
the active site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

