 |
PDBsum entry 4ukd

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.4.14
- UMP/CMP kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
CMP + ATP = CDP + ADP
|
|
2.
|
dCMP + ATP = dCDP + ADP
|
|
3.
|
UMP + ATP = UDP + ADP
|
|
 |
 |
 |
 |
 |
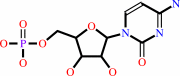 CMP
CMP
|
+
|
 ATP
ATP
|
=
|
CDP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ADP
ADP
|
|
 |
 |
 |
 |
 |
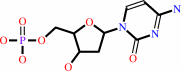 dCMP
dCMP
|
+
|
 ATP
ATP
|
=
|
dCDP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 ADP
ADP
|
|
 |
 |
 |
 |
 |
 UMP
UMP
|
+
|
 ATP
ATP
|
=
|
UDP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
ADP
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
36:9290-9296
(1997)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of active conformations of UMP kinase from Dictyostelium discoideum suggest phosphoryl transfer is associative.
|
|
I.Schlichting,
J.Reinstein.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
UMP/CMP kinase from Dictyostelium discoideum (UmpKdicty) catalyzes the specific
transfer of the terminal phosphate of ATP to UMP or CMP. Crystal structures of
UmpKdicty with substrates and the transition state analogs AlF3 or BeF2 that
lock UmpKdicty in active conformations were solved. The positions of the
catalytic Mg2+ and the highly conserved lysine of the P loop are virtually
invariant in the different structures. In contrast, catalytic arginines move to
stabilize charges that develop during this reaction. The location of the
arginines indicates formation of negative charges during the reaction at the
transferred phosphoryl group, but not at the phosphate bridging oxygen atoms.
This is consistent with an associative phosphoryl transfer mechanism but not
with a dissociative one.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.L.Whittingham,
J.Carrero-Lerida,
J.A.Brannigan,
L.M.Ruiz-Perez,
A.P.Silva,
M.J.Fogg,
A.J.Wilkinson,
I.H.Gilbert,
K.S.Wilson,
and
D.González-Pacanowska
(2010).
Structural basis for the efficient phosphorylation of AZT-MP (3'-azido-3'-deoxythymidine monophosphate) and dGMP by Plasmodium falciparum type I thymidylate kinase.
|
| |
Biochem J,
428,
499-509.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
C.Gondeau,
L.Chaloin,
P.Lallemand,
B.Roy,
C.Périgaud,
T.Barman,
A.Varga,
M.Vas,
C.Lionne,
and
S.T.Arold
(2008).
Molecular basis for the lack of enantioselectivity of human 3-phosphoglycerate kinase.
|
| |
Nucleic Acids Res,
36,
3620-3629.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.De Vivo,
M.Dal Peraro,
and
M.L.Klein
(2008).
Phosphodiester cleavage in ribonuclease H occurs via an associative two-metal-aided catalytic mechanism.
|
| |
J Am Chem Soc,
130,
10955-10962.
|
 |
|
|
|
|
 |
A.K.Hirsch,
F.R.Fischer,
and
F.Diederich
(2007).
Phosphate recognition in structural biology.
|
| |
Angew Chem Int Ed Engl,
46,
338-352.
|
 |
|
|
|
|
 |
M.De Vivo,
B.Ensing,
M.Dal Peraro,
G.A.Gomez,
D.W.Christianson,
and
M.L.Klein
(2007).
Proton shuttles and phosphatase activity in soluble epoxide hydrolase.
|
| |
J Am Chem Soc,
129,
387-394.
|
 |
|
|
|
|
 |
M.V.Dias,
L.M.Faím,
I.B.Vasconcelos,
J.S.de Oliveira,
L.A.Basso,
D.S.Santos,
and
W.F.de Azevedo
(2007).
Effects of the magnesium and chloride ions and shikimate on the structure of shikimate kinase from Mycobacterium tuberculosis.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
1-6.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.Bellinzoni,
A.Haouz,
M.Graña,
H.Munier-Lehmann,
W.Shepard,
and
P.M.Alzari
(2006).
The crystal structure of Mycobacterium tuberculosis adenylate kinase in complex with two molecules of ADP and Mg2+ supports an associative mechanism for phosphoryl transfer.
|
| |
Protein Sci,
15,
1489-1493.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Gorrell,
S.H.Lawrence,
and
J.G.Ferry
(2005).
Structural and kinetic analyses of arginine residues in the active site of the acetate kinase from Methanosarcina thermophila.
|
| |
J Biol Chem,
280,
10731-10742.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
D.Segura-Peña,
N.Sekulic,
S.Ort,
M.Konrad,
and
A.Lavie
(2004).
Substrate-induced conformational changes in human UMP/CMP kinase.
|
| |
J Biol Chem,
279,
33882-33889.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Fernandez-Fuentes,
A.Hermoso,
J.Espadaler,
E.Querol,
F.X.Aviles,
and
B.Oliva
(2004).
Classification of common functional loops of kinase super-families.
|
| |
Proteins,
56,
539-555.
|
 |
|
|
|
|
 |
R.Kagawa,
M.G.Montgomery,
K.Braig,
A.G.Leslie,
and
J.E.Walker
(2004).
The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride.
|
| |
EMBO J,
23,
2734-2744.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Z.Ahmad,
and
A.E.Senior
(2004).
Mutagenesis of residue betaArg-246 in the phosphate-binding subdomain of catalytic sites of Escherichia coli F1-ATPase.
|
| |
J Biol Chem,
279,
31505-31513.
|
 |
|
|
|
|
 |
C.Pasti,
S.Gallois-Montbrun,
H.Munier-Lehmann,
M.Veron,
A.M.Gilles,
and
D.Deville-Bonne
(2003).
Reaction of human UMP-CMP kinase with natural and analog substrates.
|
| |
Eur J Biochem,
270,
1784-1790.
|
 |
|
|
|
|
 |
L.Yu,
J.Mack,
P.J.Hajduk,
S.J.Kakavas,
A.Y.Saiki,
C.G.Lerner,
and
E.T.Olejniczak
(2003).
Solution structure and function of an essential CMP kinase of Streptococcus pneumoniae.
|
| |
Protein Sci,
12,
2613-2621.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Cook,
E.D.Lowe,
E.D.Chrysina,
V.T.Skamnaki,
N.G.Oikonomakos,
and
L.N.Johnson
(2002).
Structural studies on phospho-CDK2/cyclin A bound to nitrate, a transition state analogue: implications for the protein kinase mechanism.
|
| |
Biochemistry,
41,
7301-7311.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.C.Hutter,
and
V.Helms
(2002).
The mechanism of phosphorylation of natural nucleosides and anti-HIV analogues by nucleoside diphosphate kinase is independent of their sugar substituents.
|
| |
Chembiochem,
3,
643-651.
|
 |
|
|
|
|
 |
Madhusudan,
P.Akamine,
N.H.Xuong,
and
S.S.Taylor
(2002).
Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase.
|
| |
Nat Struct Biol,
9,
273-277.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
N.Sekulic,
L.Shuvalova,
O.Spangenberg,
M.Konrad,
and
A.Lavie
(2002).
Structural characterization of the closed conformation of mouse guanylate kinase.
|
| |
J Biol Chem,
277,
30236-30243.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
P.J.O'Brien,
and
D.Herschlag
(2002).
Alkaline phosphatase revisited: hydrolysis of alkyl phosphates.
|
| |
Biochemistry,
41,
3207-3225.
|
 |
|
|
|
|
 |
R.D.Miles,
A.Gorrell,
and
J.G.Ferry
(2002).
Evidence for a transition state analog, MgADP-aluminum fluoride-acetate, in acetate kinase from Methanosarcina thermophila.
|
| |
J Biol Chem,
277,
22547-22552.
|
 |
|
|
|
|
 |
B.Schneider,
C.Sigalat,
T.Amano,
and
J.L.Zimmermann
(2000).
Evidence for changes in the nucleotide conformation in the active site of H(+)-ATPase as determined by pulsed EPR spectroscopy.
|
| |
Biochemistry,
39,
15500-15512.
|
 |
|
|
|
|
 |
I.M.Li de La Sierra,
J.Gallay,
M.Vincent,
T.Bertrand,
P.Briozzo,
O.Bârzu,
and
A.M.Gilles
(2000).
Substrate-induced fit of the ATP binding site of cytidine monophosphate kinase from Escherichia coli: time-resolved fluorescence of 3'-anthraniloyl-2'-deoxy-ADP and molecular modeling.
|
| |
Biochemistry,
39,
15870-15878.
|
 |
|
|
|
|
 |
K.Braig,
R.I.Menz,
M.G.Montgomery,
A.G.Leslie,
and
J.E.Walker
(2000).
Structure of bovine mitochondrial F(1)-ATPase inhibited by Mg(2+) ADP and aluminium fluoride.
|
| |
Structure,
8,
567-573.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
K.Singh-Wissmann,
R.D.Miles,
C.Ingram-Smith,
and
J.G.Ferry
(2000).
Identification of essential arginines in the acetate kinase from Methanosarcina thermophila.
|
| |
Biochemistry,
39,
3671-3677.
|
 |
|
|
|
|
 |
M.C.Hutter,
and
V.Helms
(2000).
Phosphoryl transfer by a concerted reaction mechanism in UMP/CMP-kinase.
|
| |
Protein Sci,
9,
2225-2231.
|
 |
|
|
|
|
 |
N.Ostermann,
I.Schlichting,
R.Brundiers,
M.Konrad,
J.Reinstein,
T.Veit,
R.S.Goody,
and
A.Lavie
(2000).
Insights into the phosphoryltransfer mechanism of human thymidylate kinase gained from crystal structures of enzyme complexes along the reaction coordinate.
|
| |
Structure,
8,
629-642.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Nadanaciva,
J.Weber,
and
A.E.Senior
(2000).
New probes of the F1-ATPase catalytic transition state reveal that two of the three catalytic sites can assume a transition state conformation simultaneously.
|
| |
Biochemistry,
39,
9583-9590.
|
 |
|
|
|
|
 |
D.L.Graham,
J.F.Eccleston,
C.W.Chung,
and
P.N.Lowe
(1999).
Magnesium fluoride-dependent binding of small G proteins to their GTPase-activating proteins.
|
| |
Biochemistry,
38,
14981-14987.
|
 |
|
|
|
|
 |
P.Gonin,
Y.Xu,
L.Milon,
S.Dabernat,
M.Morr,
R.Kumar,
M.L.Lacombe,
J.Janin,
and
I.Lascu
(1999).
Catalytic mechanism of nucleoside diphosphate kinase investigated using nucleotide analogues, viscosity effects, and X-ray crystallography.
|
| |
Biochemistry,
38,
7265-7272.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.J.Admiraal,
B.Schneider,
P.Meyer,
J.Janin,
M.Véron,
D.Deville-Bonne,
and
D.Herschlag
(1999).
Nucleophilic activation by positioning in phosphoryl transfer catalyzed by nucleoside diphosphate kinase.
|
| |
Biochemistry,
38,
4701-4711.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
S.Nadanaciva,
J.Weber,
and
A.E.Senior
(1999).
Binding of the transition state analog MgADP-fluoroaluminate to F1-ATPase.
|
| |
J Biol Chem,
274,
7052-7058.
|
 |
|
|
|
|
 |
A.Lavie,
M.Konrad,
R.Brundiers,
R.S.Goody,
I.Schlichting,
and
J.Reinstein
(1998).
Crystal structure of yeast thymidylate kinase complexed with the bisubstrate inhibitor P1-(5'-adenosyl) P5-(5'-thymidyl) pentaphosphate (TP5A) at 2.0 A resolution: implications for catalysis and AZT activation.
|
| |
Biochemistry,
37,
3677-3686.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Lavie,
N.Ostermann,
R.Brundiers,
R.S.Goody,
J.Reinstein,
M.Konrad,
and
I.Schlichting
(1998).
Structural basis for efficient phosphorylation of 3'-azidothymidine monophosphate by Escherichia coli thymidylate kinase.
|
| |
Proc Natl Acad Sci U S A,
95,
14045-14050.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Matte,
L.W.Tari,
and
L.T.Delbaere
(1998).
How do kinases transfer phosphoryl groups?
|
| |
Structure,
6,
413-419.
|
 |
|
|
|
|
 |
H.Käck,
J.Sandmark,
K.J.Gibson,
G.Schneider,
and
Y.Lindqvist
(1998).
Crystal structure of two quaternary complexes of dethiobiotin synthetase, enzyme-MgADP-AlF3-diaminopelargonic acid and enzyme-MgADP-dethiobiotin-phosphate; implications for catalysis.
|
| |
Protein Sci,
7,
2560-2566.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.B.Berry,
and
G.N.Phillips
(1998).
Crystal structures of Bacillus stearothermophilus adenylate kinase with bound Ap5A, Mg2+ Ap5A, and Mn2+ Ap5A reveal an intermediate lid position and six coordinate octahedral geometry for bound Mg2+ and Mn2+.
|
| |
Proteins,
32,
276-288.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
P.Briozzo,
B.Golinelli-Pimpaneau,
A.M.Gilles,
J.F.Gaucher,
S.Burlacu-Miron,
H.Sakamoto,
J.Janin,
and
O.Bârzu
(1998).
Structures of escherichia coli CMP kinase alone and in complex with CDP: a new fold of the nucleoside monophosphate binding domain and insights into cytosine nucleotide specificity.
|
| |
Structure,
6,
1517-1527.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Wittinghofer
(1997).
Signaling mechanistics: aluminum fluoride for molecule of the year.
|
| |
Curr Biol,
7,
R682-R685.
|
 |
|
|
|
|
 |
M.R.Ahmadian,
P.Stege,
K.Scheffzek,
and
A.Wittinghofer
(1997).
Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras.
|
| |
Nat Struct Biol,
4,
686-689.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

