 |
PDBsum entry 4rv8

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
4rv8
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Co-crystal structure of the catalytic domain of the inosine monophosphate dehydrogenase from cryptosporidium parvum and the inhibitor p131
|
|
Structure:
|
 |
Inosine-5'-monophosphate dehydrogenase. Chain: a, b, c, d. Fragment: catalytic domain of impdh. Synonym: imp dehydrogenase, impd, impdh. Engineered: yes. Mutation: yes
|
|
Source:
|
 |
Cryptosporidium parvum. Organism_taxid: 5807. Gene: 56k.02, cgd6_20. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.05Å
|
R-factor:
|
0.189
|
R-free:
|
0.229
|
|
|
Authors:
|
 |
Y.Kim,M.Makowska-Grzyska,M.Gu,M.Kavitha,L.Hedstrom,W.F.Anderson, A.Joachimiak,Center For Structural Genomics Of Infectious Diseases (Csgid)
|
|
Key ref:
|
 |
Y.Kim
et al.
(2015).
Structure of Cryptosporidium IMP dehydrogenase bound to an inhibitor with in vivo antiparasitic activity.
Acta Crystallogr F Struct Biol Commun,
71,
531-538.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
25-Nov-14
|
Release date:
|
31-Dec-14
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8T6T2
(IMDH_CRYPV) -
Inosine-5'-monophosphate dehydrogenase from Cryptosporidium parvum
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
400 a.a.
327 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 3 residue positions (black
crosses)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.205
- Imp dehydrogenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP and GMP Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
IMP + NAD+ + H2O = XMP + NADH + H+
|
 |
 |
 |
 |
 |
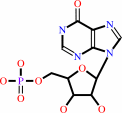 IMP
IMP
|
+
|
NAD(+)
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
=
|
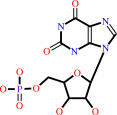 XMP
XMP
|
+
|
 NADH
NADH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr F Struct Biol Commun
71:531-538
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of Cryptosporidium IMP dehydrogenase bound to an inhibitor with in vivo antiparasitic activity.
|
|
Y.Kim,
M.Makowska-Grzyska,
S.K.Gorla,
D.R.Gollapalli,
G.D.Cuny,
A.Joachimiak,
L.Hedstrom.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Inosine 5'-monophosphate dehydrogenase (IMPDH) is a promising target for the
treatment of Cryptosporidium infections. Here, the structure of C. parvum IMPDH
(CpIMPDH) in complex with inosine 5'-monophosphate (IMP) and P131, an inhibitor
with in vivo anticryptosporidial activity, is reported. P131 contains two
aromatic groups, one of which interacts with the hypoxanthine ring of IMP, while
the second interacts with the aromatic ring of a tyrosine in the adjacent
subunit. In addition, the amine and NO2 moieties bind in hydrated cavities,
forming water-mediated hydrogen bonds to the protein. The design of compounds to
replace these water molecules is a new strategy for the further optimization of
C. parvum inhibitors for both antiparasitic and antibacterial applications.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

