 |
PDBsum entry 4oqc

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxygen binding
|
PDB id
|
|

|

|
4oqc
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.7.3.3
- factor independent urate hydroxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
AMP Catabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
urate + O2 + H2O = 5-hydroxyisourate + H2O2
|
 |
 |
 |
 |
 |
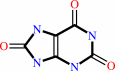 urate
urate
|
+
|
 O2
O2
|
+
|
H2O
|
=
|
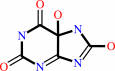 5-hydroxyisourate
5-hydroxyisourate
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Copper
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallogr F Struct Biol Commun
70:896-902
(2014)
|
|
PubMed id:
|
|
|
|

|
| |
|
Azide inhibition of urate oxidase.
|
|
L.Gabison,
N.Colloc'h,
T.Prangé.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The inhibition of urate oxidase (UOX) by azide was investigated by X-ray
diffraction techniques and compared with cyanide inhibition. Two well
characterized sites for reagents are present in the enzyme: the dioxygen site
and the substrate-binding site. To examine the selectivity of these sites
towards azide inhibition, several crystallization conditions were developed. UOX
was co-crystallized with azide (N3) in the presence or absence of either uric
acid (UA, the natural substrate) or 8-azaxanthine (8AZA, a competitive
inhibitor). In a second set of experiments, previously grown orthorhombic
crystals of the UOX-UA or UOX-8AZA complexes were soaked in sodium azide
solutions. In a third set of experiments, orthorhombic crystals of UOX with the
exchangeable ligand 8-nitroxanthine (8NXN) were soaked in a solution containing
uric acid and azide simultaneously (competitive soaking). In all assays, the
soaking periods were either short (a few hours) or long (one or two months).
These different experimental conditions showed that one or other of the sites,
or the two sites together, could be inhibited. This also demonstrated that azide
not only competes with dioxygen as cyanide does but also competes with the
substrate for its enzymatic site. A model in agreement with experimental data
would be an azide in equilibrium between two sites, kinetically in favour of the
dioxygen site and thermodynamically in favour of the substrate-binding site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

