 |
PDBsum entry 4oeh
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
X-ray structure of uridine phosphorylase from vibrio cholerae complexed with uracil at 1.91 a resolution
|
|
Structure:
|
 |
Uridine phosphorylase. Chain: a, b, c, d, e, f. Engineered: yes
|
|
Source:
|
 |
Vibrio cholerae o1 biovar el tor. Organism_taxid: 243277. Strain: 569b. Gene: udp, vc_1034. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.91Å
|
R-factor:
|
0.174
|
R-free:
|
0.216
|
|
|
Authors:
|
 |
I.I.Prokofev,A.A.Lashkov,A.G.Gabdoulkhakov,C.Betzel,A.M.Mikhailov
|
|
Key ref:
|
 |
I.I.Prokofev
et al.
X-Ray structure of uridine phosphorylase from vibrio cholerae complexed with uracil at 1.91 a resolution.
To be published,
.
|
 |
|
Date:
|
 |
|
13-Jan-14
|
Release date:
|
04-Mar-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q9K4U1
(Q9K4U1_VIBCL) -
Uridine phosphorylase from Vibrio cholerae
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
253 a.a.
251 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.4.2.3
- uridine phosphorylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
uridine + phosphate = alpha-D-ribose 1-phosphate + uracil
|
 |
 |
 |
 |
 |
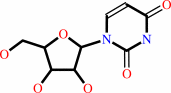 uridine
uridine
|
+
|
 phosphate
phosphate
|
=
|
alpha-D-ribose 1-phosphate
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
uracil
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

