 |
PDBsum entry 4mv6
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Crystal structure of biotin carboxylase from haemophilus influenzae in complex with phosphonoacetamide
|
|
Structure:
|
 |
Biotin carboxylase. Chain: a. Synonym: acetyl-coa carboxylase subunit a, acc. Engineered: yes
|
|
Source:
|
 |
Haemophilus influenzae. Organism_taxid: 71421. Strain: atcc 51907 / dsm 11121 / kw20 / rd. Gene: accc, hi_0972. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.77Å
|
R-factor:
|
0.176
|
R-free:
|
0.203
|
|
|
Authors:
|
 |
T.C.Broussard,S.Pakhomova,D.B.Neau,T.S.Champion,R.J.Bonnot, G.L.Waldrop
|
|
Key ref:
|
 |
T.C.Broussard
et al.
(2015).
Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
Biochemistry,
54,
3860-3870.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
23-Sep-13
|
Release date:
|
14-Jan-15
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P43873
(ACCC_HAEIN) -
Biotin carboxylase from Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
448 a.a.
412 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 1:
|
 |
E.C.6.3.4.14
- biotin carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N6-biotinyl-L-lysyl-[protein] + hydrogencarbonate + ATP = N6- carboxybiotinyl-L-lysyl-[protein] + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
N(6)-biotinyl-L-lysyl-[protein]
|
+
|
 hydrogencarbonate
hydrogencarbonate
|
+
|
 ATP
ATP
|
=
|
N(6)- carboxybiotinyl-L-lysyl-[protein]
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 44.44% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.6.4.1.2
- acetyl-CoA carboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
hydrogencarbonate + acetyl-CoA + ATP = malonyl-CoA + ADP + phosphate + H+
|
 |
 |
 |
 |
 |
 hydrogencarbonate
hydrogencarbonate
|
+
|
 acetyl-CoA
acetyl-CoA
|
+
|
 ATP
ATP
|
=
|
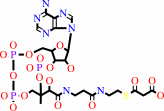 malonyl-CoA
malonyl-CoA
|
+
|
 ADP
ADP
|
+
|
 phosphate
phosphate
|
+
|
H(+)
Bound ligand (Het Group name = )
matches with 44.44% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Biotin
|
 |
 |
 |
 |
 |
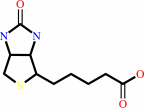 Biotin
Biotin
|
|
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
54:3860-3870
(2015)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural Analysis of Substrate, Reaction Intermediate, and Product Binding in Haemophilus influenzae Biotin Carboxylase.
|
|
T.C.Broussard,
S.Pakhomova,
D.B.Neau,
R.Bonnot,
G.L.Waldrop.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Acetyl-CoA carboxylase catalyzes the first and regulated step in fatty acid
synthesis. In most Gram-negative and Gram-positive bacteria, the enzyme is
composed of three proteins: biotin carboxylase, a biotin carboxyl carrier
protein (BCCP), and carboxyltransferase. The reaction mechanism involves two
half-reactions with biotin carboxylase catalyzing the ATP-dependent
carboxylation of biotin-BCCP in the first reaction. In the second reaction,
carboxyltransferase catalyzes the transfer of the carboxyl group from
biotin-BCCP to acetyl-CoA to form malonyl-CoA. In this report, high-resolution
crystal structures of biotin carboxylase from Haemophilus influenzae were
determined with bicarbonate, the ATP analogue AMPPCP; the carboxyphosphate
intermediate analogues, phosphonoacetamide and phosphonoformate; the products
ADP and phosphate; and the carboxybiotin analogue N1'-methoxycarbonyl biotin
methyl ester. The structures have a common theme in that bicarbonate, phosphate,
and the methyl ester of the carboxyl group of N1'-methoxycarbonyl biotin methyl
ester all bound in the same pocket in the active site of biotin carboxylase and
as such utilize the same set of amino acids for binding. This finding suggests a
catalytic mechanism for biotin carboxylase in which the binding pocket that
binds tetrahedral phosphate also accommodates and stabilizes a tetrahedral
dianionic transition state resulting from direct transfer of CO2 from the
carboxyphosphate intermediate to biotin.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

