 |
PDBsum entry 4jax
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of dimeric klhxk1 in crystal form x
|
|
Structure:
|
 |
Hexokinase. Chain: a, b, c, d, e, f. Engineered: yes
|
|
Source:
|
 |
Kluyveromyces lactis. Yeast. Organism_taxid: 284590. Strain: cbs2359/152. Gene: klla0d11352g, rag5. Expressed in: kluyveromyces lactis. Expression_system_taxid: 28985.
|
|
Resolution:
|
 |
|
2.26Å
|
R-factor:
|
0.201
|
R-free:
|
0.240
|
|
|
Authors:
|
 |
E.B.Kuettner,N.Strater,K.Kettner,A.Otto,H.Lilie,R.P.Golbik, T.M.Kriegel
|
|
Key ref:
|
 |
K.Kettner
et al.
(2013).
In vivo phosphorylation and in vitro autophosphorylation-inactivation of Kluyveromyces lactis hexokinase KlHxk1.
Biochem Biophys Res Commun,
435,
313-318.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
19-Feb-13
|
Release date:
|
01-May-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P33284
(HXK_KLULA) -
Hexokinase from Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
485 a.a.
470 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.1.1
- hexokinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a D-hexose + ATP = a D-hexose 6-phosphate + ADP + H+
|
 |
 |
 |
 |
 |
D-hexose
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
 ATP
ATP
|
=
|
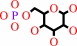 D-hexose 6-phosphate
D-hexose 6-phosphate
|
+
|
 ADP
ADP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochem Biophys Res Commun
435:313-318
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
In vivo phosphorylation and in vitro autophosphorylation-inactivation of Kluyveromyces lactis hexokinase KlHxk1.
|
|
K.Kettner,
E.B.Kuettner,
A.Otto,
H.Lilie,
R.P.Golbik,
N.Sträter,
T.M.Kriegel.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The bifunctional hexokinase KlHxk1 is a key component of glucose-dependent
signal transduction in Kluyveromyces lactis. KlHxk1 is phosphorylated in vivo
and undergoes ATP-dependent autophosphorylation-inactivation in vitro. This
study identifies serine-15 as the site of in vivo phosphorylation and serine-157
as the autophosphorylation-inactivation site. X-ray crystallography of the in
vivo phosphorylated enzyme indicates the existence of a ring-shaped symmetrical
homodimer carrying two phosphoserine-15 residues. In contrast, small-angle X-ray
scattering and equilibrium sedimentation analyses reveal the existence of
monomeric phosphoserine-15 KlHxk1 in solution. While phosphorylation at
serine-15 and concomitant homodimer dissociation are likely to be involved in
glucose signalling, mechanism and putative physiological significance of KlHxk1
inactivation by autophosphorylation at serine-157 remain to be established.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

