 |
PDBsum entry 4i4c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Unknown function
|
PDB id
|
|

|

|
4i4c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Unknown function
|
 |
|
Title:
|
 |
Crystal structure of the protein frsa complexed with unknown ligand
|
|
Structure:
|
 |
Upf0255 protein frsa. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Vibrio vulnificus. Organism_taxid: 672. Gene: frsa. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
1.95Å
|
R-factor:
|
0.182
|
R-free:
|
0.227
|
|
|
Authors:
|
 |
A.A.Fedorov,E.V.Fedorov,B.Desai,J.A.Gerlt,N.Richards,S.C.Almo
|
|
Key ref:
|
 |
W.F.Kellett
et al.
(2013).
Computational, structural, and kinetic evidence that Vibrio vulnificus FrsA is not a cofactor-independent pyruvate decarboxylase.
Biochemistry,
52,
1842-1844.
PubMed id:
DOI:
|
 |
|
Date:
|
 |
|
27-Nov-12
|
Release date:
|
09-Oct-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
D9IR22
(D9IR22_VIBVL) -
Esterase FrsA from Vibrio vulnificus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
415 a.a.
402 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.1.1
- carboxylesterase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
a carboxylic ester + H2O = an alcohol + a carboxylate + H+
|
 |
 |
 |
 |
 |
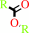 carboxylic ester
carboxylic ester
|
+
|
H2O
|
=
|
 alcohol
alcohol
|
+
|
 carboxylate
carboxylate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
52:1842-1844
(2013)
|
|
PubMed id:
|
|
|
|

|
| |
|
Computational, structural, and kinetic evidence that Vibrio vulnificus FrsA is not a cofactor-independent pyruvate decarboxylase.
|
|
W.F.Kellett,
E.Brunk,
B.J.Desai,
A.A.Fedorov,
S.C.Almo,
J.A.Gerlt,
U.Rothlisberger,
N.G.Richards.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The fermentation-respiration switch (FrsA) protein in Vibrio vulnificus was
recently reported to catalyze the cofactor-independent decarboxylation of
pyruvate. We now report quantum mechanical/molecular mechenical calculations
that examine the energetics of C-C bond cleavage for a pyruvate molecule bound
within the putative active site of FrsA. These calculations suggest that the
barrier to C-C bond cleavage in the bound substrate is 28 kcal/mol, which is
similar to that estimated for the uncatalyzed decarboxylation of pyruvate in
water at 25 °C. In agreement with the theoretical predictions, no pyruvate
decarboxylase activity was detected for recombinant FrsA protein that could be
crystallized and structurally characterized. These results suggest that the
functional annotation of FrsA as a cofactor-independent pyruvate decarboxylase
is incorrect.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

