 |
PDBsum entry 4efc
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of adenylosuccinate lyase from trypanosoma brucei, tb427tmp.160.5560
|
|
Structure:
|
 |
Adenylosuccinate lyase. Chain: a, b. Engineered: yes
|
|
Source:
|
 |
Trypanosoma brucei brucei. Organism_taxid: 999953. Strain: 927/4 gutat10.1. Gene: tb09.160.5560. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.00Å
|
R-factor:
|
0.151
|
R-free:
|
0.191
|
|
|
Authors:
|
 |
A.K.Wernimont,P.Loppnau,K.T.Osman,C.H.Arrowsmith,A.M.Edwards, C.Bountra,D.A.Robinson,P.G.Wyatt,I.H.Gilbert,A.H.Fairlamb,R.Hui, Y.H.Lin,Structural Genomics Consortium (Sgc)
|
|
Key ref:
|
 |
A.K.Wernimont
et al.
Crystal structure of adenylosuccinate lyase from tryp brucei, Tb427tmp.160.5560.
To be published,
.
|
 |
|
Date:
|
 |
|
29-Mar-12
|
Release date:
|
13-Feb-13
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q38EJ2
(Q38EJ2_TRYB2) -
Adenylosuccinate lyase from Trypanosoma brucei brucei (strain 927/4 GUTat10.1)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
471 a.a.
452 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.3.2.2
- adenylosuccinate lyase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Purine Biosynthesis (late stages)
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
N6-(1,2-dicarboxyethyl)-AMP = fumarate + AMP
|
|
2.
|
(2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamido]succinate = 5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamide + fumarate
|
|
 |
 |
 |
 |
 |
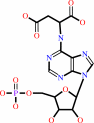 N(6)-(1,2-dicarboxyethyl)-AMP
N(6)-(1,2-dicarboxyethyl)-AMP
|
=
|
fumarate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 AMP
AMP
|
|
 |
 |
 |
 |
 |
(2S)-2-[5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamido]succinate
|
=
|
5-amino-1-(5-phospho-beta-D-ribosyl)imidazole-4- carboxamide
|
+
|
 fumarate
fumarate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

