 |
PDBsum entry 3v75
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of putative orotidine 5'-phosphate decarboxylase from streptomyces avermitilis ma-4680
|
|
Structure:
|
 |
Orotidine 5'-phosphate decarboxylase. Chain: a. Synonym: omp decarboxylase. Engineered: yes
|
|
Source:
|
 |
Streptomyces avermitilis. Organism_taxid: 33903. Strain: ma-4680. Gene: pyrf, sav6869, sav_6869. Expressed in: escherichia coli. Expression_system_taxid: 511693.
|
|
Resolution:
|
 |
|
1.40Å
|
R-factor:
|
0.149
|
R-free:
|
0.172
|
|
|
Authors:
|
 |
P.J.Stogios,X.Xu,H.Cui,M.Kudritska,K.Tan,A.Edwards,A.Savchenko, A.Joachimiak,Midwest Center For Structural Genomics (Mcsg)
|
|
Key ref:
|
 |
P.J.Stogios
et al.
Crystal structure of putative orotidine 5'-Phosphate decarboxylase from streptomyces avermitilis ma-4680.
To be published,
.
|
 |
|
Date:
|
 |
|
20-Dec-11
|
Release date:
|
09-May-12
|
|
|
Supersedes:
|

|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q827Q5
(Q827Q5_STRAW) -
Orotidine 5'-phosphate decarboxylase from Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
280 a.a.
285 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
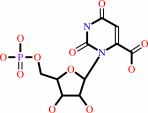 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
|
=
|
 UMP
UMP
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

