 |
PDBsum entry 3k4c

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3k4c
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.3.10
- pyranose oxidase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-glucose + O2 = 2-dehydro-D-glucose + H2O2
|
 |
 |
 |
 |
 |
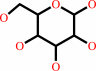 D-glucose
D-glucose
|
+
|
 O2
O2
|
=
|
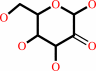 2-dehydro-D-glucose
2-dehydro-D-glucose
|
+
|
 H2O2
H2O2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
FAD
Bound ligand (Het Group name =
FAD)
corresponds exactly
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Biol Chem
285:9697-9705
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
A conserved active-site threonine is important for both sugar and flavin oxidations of pyranose 2-oxidase.
|
|
W.Pitsawong,
J.Sucharitakul,
M.Prongjit,
T.C.Tan,
O.Spadiut,
D.Haltrich,
C.Divne,
P.Chaiyen.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Pyranose 2-oxidase (P2O) catalyzes the oxidation by O(2) of d-glucose and
several aldopyranoses to yield the 2-ketoaldoses and H(2)O(2). Based on crystal
structures, in one rotamer conformation, the threonine hydroxyl of Thr(169)
forms H-bonds to the flavin-N5/O4 locus, whereas, in a different rotamer, it may
interact with either sugar or other parts of the P2O.sugar complex. Transient
kinetics of wild-type (WT) and Thr(169) --> S/N/G/A replacement variants show
that D-Glc binds to T169S, T169N, and WT with the same K(d) (45-47 mm), and the
hydride transfer rate constants (k(red)) are similar (15.3-9.7 s(-1) at 4
degrees C). k(red) of T169G with D-glucose (0.7 s(-1), 4 degrees C) is
significantly less than that of WT but not as severely affected as in T169A
(k(red) of 0.03 s(-1) at 25 degrees C). Transient kinetics of WT and mutants
using d-galactose show that P2O binds d-galactose with a one-step binding
process, different from binding of d-glucose. In T169S, T169N, and T169G, the
overall turnover with d-Gal is faster than that of WT due to an increase of
k(red). In the crystal structure of T169S, Ser(169) O gamma assumes a position
identical to that of O gamma 1 in Thr(169); in T169G, solvent molecules may be
able to rescue H-bonding. Our data suggest that a competent reductive
half-reaction requires a side chain at position 169 that is able to form an
H-bond within the ES complex. During the oxidative half-reaction, all mutants
failed to stabilize a C4a-hydroperoxyflavin intermediate, thus suggesting that
the precise position and geometry of the Thr(169) side chain are required for
intermediate stabilization.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Salaheddin,
Y.Takakura,
M.Tsunashima,
B.Stranzinger,
O.Spadiut,
M.Yamabhai,
C.K.Peterbauer,
and
D.Haltrich
(2010).
Characterisation of recombinant pyranose oxidase from the cultivated mycorrhizal basidiomycete Lyophyllum shimeji (hon-shimeji).
|
| |
Microb Cell Fact,
9,
57.
|
 |
|
|
|
|
 |
O.Spadiut,
T.C.Tan,
I.Pisanelli,
D.Haltrich,
and
C.Divne
(2010).
Importance of the gating segment in the substrate-recognition loop of pyranose 2-oxidase.
|
| |
FEBS J,
277,
2892-2909.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

