 |
PDBsum entry 3i1c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.1.8.2
- Transferred entry: 3.8.2.2.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
diisopropyl fluorophosphate + H2O = diisopropyl phosphate + fluoride + 2 H+
|
 |
 |
 |
 |
 |
 diisopropyl fluorophosphate
diisopropyl fluorophosphate
|
+
|
H2O
|
=
|
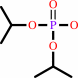 diisopropyl phosphate
diisopropyl phosphate
|
+
|
fluoride
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Science
329:309-313
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Computational design of an enzyme catalyst for a stereoselective bimolecular Diels-Alder reaction.
|
|
J.B.Siegel,
A.Zanghellini,
H.M.Lovick,
G.Kiss,
A.R.Lambert,
J.L.St Clair,
J.L.Gallaher,
D.Hilvert,
M.H.Gelb,
B.L.Stoddard,
K.N.Houk,
F.E.Michael,
D.Baker.
|
|
|

|
| |
ABSTRACT
|
|

|
| |
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
N.Koga,
R.Tatsumi-Koga,
G.Liu,
R.Xiao,
T.B.Acton,
G.T.Montelione,
and
D.Baker
(2012).
Principles for designing ideal protein structures.
|
| |
Nature,
491,
222-227.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
U.T.Bornscheuer,
G.W.Huisman,
R.J.Kazlauskas,
S.Lutz,
J.C.Moore,
and
K.Robins
(2012).
Engineering the third wave of biocatalysis.
|
| |
Nature,
485,
185-194.
|
 |
|
|
|
|
 |
A.Morin,
K.W.Kaufmann,
C.Fortenberry,
J.M.Harp,
L.S.Mizoue,
and
J.Meiler
(2011).
Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
|
| |
Protein Eng Des Sel,
24,
503-516.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.S.Bommarius,
J.K.Blum,
and
M.J.Abrahamson
(2011).
Status of protein engineering for biocatalysts: how to design an industrially useful biocatalyst.
|
| |
Curr Opin Chem Biol,
15,
194-200.
|
 |
|
|
|
|
 |
B.M.Nestl,
B.A.Nebel,
and
B.Hauer
(2011).
Recent progress in industrial biocatalysis.
|
| |
Curr Opin Chem Biol,
15,
187-193.
|
 |
|
|
|
|
 |
E.M.Brustad,
and
F.H.Arnold
(2011).
Optimizing non-natural protein function with directed evolution.
|
| |
Curr Opin Chem Biol,
15,
201-210.
|
 |
|
|
|
|
 |
H.S.Rzepa
(2011).
Can 1,3-dimethylcyclobutadiene and carbon dioxide co-exist inside a supramolecular cavity?
|
| |
Chem Commun (Camb),
47,
1851-1853.
|
 |
|
|
|
|
 |
J.T.MacDonald,
C.Barnes,
R.I.Kitney,
P.S.Freemont,
and
G.B.Stan
(2011).
Computational design approaches and tools for synthetic biology.
|
| |
Integr Biol (Camb),
3,
97.
|
 |
|
|
|
|
 |
M.M.Stratton,
and
S.N.Loh
(2011).
Converting a protein into a switch for biosensing and functional regulation.
|
| |
Protein Sci,
20,
19-29.
|
 |
|
|
|
|
 |
P.J.Deuss,
R.den Heeten,
W.Laan,
and
P.C.Kamer
(2011).
Bioinspired catalyst design and artificial metalloenzymes.
|
| |
Chemistry,
17,
4680-4698.
|
 |
|
|
|
|
 |
Y.Mo,
P.Bao,
and
J.Gao
(2011).
Energy decomposition analysis based on a block-localized wavefunction and multistate density functional theory.
|
| |
Phys Chem Chem Phys,
13,
6760-6775.
|
 |
|
|
|
|
 |
A.Doerr
(2010).
Molecular engineering: Unnatural design.
|
| |
Nat Methods,
7,
671.
|
 |
|
|
|
|
 |
B.R.Fritz,
L.E.Timmerman,
N.M.Daringer,
J.N.Leonard,
and
M.C.Jewett
(2010).
Biology by design: from top to bottom and back.
|
| |
J Biomed Biotechnol,
2010,
232016.
|
 |
|
|
|
|
 |
C.Jäckel,
and
D.Hilvert
(2010).
Biocatalysts by evolution.
|
| |
Curr Opin Biotechnol,
21,
753-759.
|
 |
|
|
|
|
 |
D.Baker
(2010).
An exciting but challenging road ahead for computational enzyme design.
|
| |
Protein Sci,
19,
1817-1819.
|
 |
|
|
|
|
 |
G.Kiss,
D.Röthlisberger,
D.Baker,
and
K.N.Houk
(2010).
Evaluation and ranking of enzyme designs.
|
| |
Protein Sci,
19,
1760-1773.
|
 |
|
|
|
|
 |
J.D.Keasling
(2010).
Manufacturing molecules through metabolic engineering.
|
| |
Science,
330,
1355-1358.
|
 |
|
|
|
|
 |
M.Harmata
(2010).
The (4+3)-cycloaddition reaction: simple allylic cations as dienophiles.
|
| |
Chem Commun (Camb),
46,
8886-8903.
|
 |
|
|
|
|
 |
S.Lutz
(2010).
Biochemistry. Reengineering enzymes.
|
| |
Science,
329,
285-287.
|
 |
|
|
|
|
 |
S.Lutz
(2010).
Beyond directed evolution--semi-rational protein engineering and design.
|
| |
Curr Opin Biotechnol,
21,
734-743.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

