 |
PDBsum entry 3hhs

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3hhs
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B:
E.C.1.14.18.1
- tyrosinase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Melanin Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
L-tyrosine + O2 = L-dopaquinone + H2O
|
|
2.
|
2 L-dopa + O2 = 2 L-dopaquinone + 2 H2O
|
|
 |
 |
 |
 |
 |
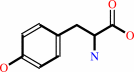 L-tyrosine
L-tyrosine
|
+
|
 O2
O2
|
=
|
L-dopaquinone
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
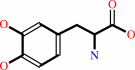 2
×
L-dopa
2
×
L-dopa
|
+
|
 O2
O2
|
=
|
2
×
L-dopaquinone
|
+
|
2
×
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Cu cation
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
106:17002-17006
(2009)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of Manduca sexta prophenoloxidase provides insights into the mechanism of type 3 copper enzymes.
|
|
Y.Li,
Y.Wang,
H.Jiang,
J.Deng.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Arthropod phenoloxidase (PO) generates quinones and other toxic compounds to
sequester and kill pathogens during innate immune responses. It is also involved
in wound healing and other physiological processes. Insect PO is activated from
its inactive precursor, prophenoloxidase (PPO), by specific proteolysis via a
serine protease cascade. Here, we report the crystal structure of PPO from a
lepidopteran insect at a resolution of 1.97 A, which is the initial structure
for a PPO from the type 3 copper protein family. Manduca sexta PPO is a
heterodimer consisting of 2 homologous polypeptide chains, PPO1 and PPO2. The
active site of each subunit contains a canonical type 3 di-nuclear copper
center, with each copper ion coordinated with 3 structurally conserved
histidines. The acidic residue Glu-395 located at the active site of PPO2 may
serve as a general base for deprotonation of monophenolic substrates, which is
key to the ortho-hydroxylase activity of PO. The structure provides unique
insights into the mechanism by which type 3 copper proteins differ in their
enzymatic activities, albeit sharing a common active center. A drastic change in
electrostatic surface induced on cleavage at Arg-51 allows us to propose a model
for localized PPO activation in insects.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Overall structure of M. sexta PPO. (A) The heterodimeric PPO
is formed in a back-to-back mode. PPO1 and PPO2 are shown in
green and yellow, respectively. (B) Domains of PPO2 are colored
as follows: pro-region, purple; domain I, blue; domain II,
yellow; domain III, green. The di-copper atoms are located in
domain II and are shown as red spheres. The proteolytic site R51
residue is shown in the stick. The amino-terminus and
carboxyl-terminus of PPO2 are indicated as N and C, respectively.
|
 |
Figure 2.
Di-copper center in M. sexta PPO2. The active site of PPO2
can be superimposed well with that of oxygenated Limulus
polyphemus hemocyanin (Lp-HC, PDB ID code 1OXY). The secondary
structures of PPO2 are shown in the ribbon and colored in
yellow. The 6 copper-coordinating His ligands are shown as
sticks, with those from Lp-HC colored green. The di-copper atoms
are shown as spheres: PPO2, red; Lp-HC, purple. The peroxide ion
in Lp-HC is shown as brown spheres. Notice the unique E395 in
PPO2, which is located near the substrate place holder F88. E395
could be a base for phenol deprotonation, which is key to the
ortho-phenol hydroxylation activity of PPO.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
Y.Kawamura-Konishi,
S.Maekawa,
M.Tsuji,
and
H.Goto
(2011).
C-terminal processing of tyrosinase is responsible for activation of Pholiota microspora proenzyme.
|
| |
Appl Microbiol Biotechnol,
90,
227-234.
|
 |
|
|
|
|
 |
Y.X.Si,
S.J.Yin,
D.Park,
H.Y.Chung,
L.Yan,
Z.R.Lü,
H.M.Zhou,
J.M.Yang,
G.Y.Qian,
and
Y.D.Park
(2011).
Tyrosinase inhibition by isophthalic acid: kinetics and computational simulation.
|
| |
Int J Biol Macromol,
48,
700-704.
|
 |
|
|
|
|
 |
L.Cerenius,
S.Kawabata,
B.L.Lee,
M.Nonaka,
and
K.Söderhäll
(2010).
Proteolytic cascades and their involvement in invertebrate immunity.
|
| |
Trends Biochem Sci,
35,
575-583.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
|

