 |
PDBsum entry 3eoo

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
2.9a crystal structure of methyl-isocitrate lyase from burkholderia pseudomallei
|
|
Structure:
|
 |
Methylisocitrate lyase. Chain: a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p. Engineered: yes
|
|
Source:
|
 |
Burkholderia pseudomallei 1655. Organism_taxid: 331109. Strain: 1710b. Gene: burps1655_c0444, burps1710b_2458, burps1710b_3237, msrb, prpb. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.90Å
|
R-factor:
|
0.240
|
R-free:
|
0.299
|
|
|
Authors:
|
 |
Seattle Structural Genomics Center For Infectious Disease (Ssgcid)
|
|
Key ref:
|
 |
Ssgcid
Crystal structures of isocitrate lyase from brucella melitensis.
To be published,
.
|
 |
|
Date:
|
 |
|
28-Sep-08
|
Release date:
|
21-Oct-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.3.30
- methylisocitrate lyase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate = pyruvate + succinate
|
 |
 |
 |
 |
 |
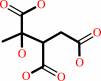 (2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate
(2S,3R)-3-hydroxybutane-1,2,3-tricarboxylate
|
=
|
 pyruvate
pyruvate
|
+
|
 succinate
succinate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

