 |
PDBsum entry 3cli

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of arabidopsis thaliana allene oxide synthase (aos, cytochrome p450 74a, cyp74a) at 1.80 a resolution
|
|
Structure:
|
 |
Allene oxide synthase. Chain: a, b. Synonym: cytochrome p450 74a, chloroplast. Hydroperoxide dehydrase. Engineered: yes
|
|
Source:
|
 |
Arabidopsis thaliana. Mouse-ear cress. Organism_taxid: 3702. Gene: aos, cyp74a. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.80Å
|
R-factor:
|
0.171
|
R-free:
|
0.198
|
|
|
Authors:
|
 |
D.-S.Lee,P.Nioche,C.S.Raman
|
Key ref:

|
 |
D.S.Lee
et al.
(2008).
Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes.
Nature,
455,
363-368.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
19-Mar-08
|
Release date:
|
19-Aug-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q96242
(CP74A_ARATH) -
Allene oxide synthase, chloroplastic from Arabidopsis thaliana
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
518 a.a.
465 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.2.1.92
- hydroperoxide dehydratase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
(13S)-hydroperoxy-(9Z,11E,15Z)-octadecatrienoate = (9Z,13S,15Z)-12,13- epoxyoctadeca-9,11,15-trienoate + H2O
|
 |
 |
 |
 |
 |
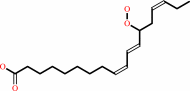 (9Z,11E,15Z)-(13S)-hydroperoxyoctadeca-9,11,15-trienoate
(9Z,11E,15Z)-(13S)-hydroperoxyoctadeca-9,11,15-trienoate
|
=
|
(9Z,15Z)- (13S)-12,13-epoxyoctadeca-9,11,15-trienoate
|
+
|
H(2)O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme-thiolate
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Nature
455:363-368
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insights into the evolutionary paths of oxylipin biosynthetic enzymes.
|
|
D.S.Lee,
P.Nioche,
M.Hamberg,
C.S.Raman.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The oxylipin pathway generates not only prostaglandin-like jasmonates but also
green leaf volatiles (GLVs), which confer characteristic aromas to fruits and
vegetables. Although allene oxide synthase (AOS) and hydroperoxide lyase are
atypical cytochrome P450 family members involved in the synthesis of jasmonates
and GLVs, respectively, it is unknown how these enzymes rearrange their
hydroperoxide substrates into different products. Here we present the crystal
structures of Arabidopsis thaliana AOS, free and in complex with substrate or
intermediate analogues. The structures reveal an unusual active site poised to
control the reactivity of an epoxyallylic radical and its cation by means of
interactions with an aromatic pi-system. Replacing the amino acid involved in
these steps by a non-polar residue markedly reduces AOS activity and,
unexpectedly, is both necessary and sufficient for converting AOS into a GLV
biosynthetic enzyme. Furthermore, by combining our structural data with
bioinformatic and biochemical analyses, we have discovered previously unknown
hydroperoxide lyase in plant growth-promoting rhizobacteria, AOS in coral, and
epoxyalcohol synthase in amphioxus. These results indicate that oxylipin
biosynthetic genes were present in the last common ancestor of plants and
animals, but were subsequently lost in all metazoan lineages except Placozoa,
Cnidaria and Cephalochordata.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1: Reactions catalysed by the CYP74 enzyme family. In
higher plants, C[18] fatty acids (linoleic and linolenic acids)
are oxygenated at either position 9 or 13 by lipoxygenases to
yield hydroperoxides. Subsequently, these are converted by
allene oxide synthase (AOS, also known as CYP74A), hydroperoxide
lyase (HPL, also known as CYP74B) and divinyl ether synthase
(DES, also known as CYP74D) to allene oxide (an essential
intermediate in jasmonate biosynthesis), green leaf volatiles
(aldehydes) and divinyl ethers, respectively. For clarity, only
13-hydroperoxide-derived metabolites are shown. 12,13(S)-allene
oxide, 12,13S-epoxy-9Z,11,15Z-octadecatrienoic acid; 13(S)-HPOT,
13S-hydroperoxyoctadecatrienoic acid;  -ketol
is the hydrolytic product of the highly unstable allene oxide;
hemiacetal, hydroxyhexenyloxydodecadienoic acid. -ketol
is the hydrolytic product of the highly unstable allene oxide;
hemiacetal, hydroxyhexenyloxydodecadienoic acid.
|
 |
Figure 4.
Figure 4: Proposed reaction paths for AOS and HPL on the basis
of the current structural and enzymological studies^14, ^15,
^16, ^17, ^21. The intermediate epoxyallylic radical formed
in step 4 can either undergo one electron oxidation followed by
proton loss (AOS) or oxygen rebound (HPL). The structure of the
peroxide substrate is abbreviated to highlight the region
undergoing chemical transformation. For clarity, the Fe–S bond
between the haem iron and Cys 471 is only shown in step 1. It
remains intact throughout the catalytic cycle. Hydrogen bonds
are illustrated with blue dashed lines.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Macmillan Publishers Ltd:
Nature
(2008,
455,
363-368)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Nelson,
and
D.Werck-Reichhart
(2011).
A P450-centric view of plant evolution.
|
| |
Plant J,
66,
194-211.
|
 |
|
|
|
|
 |
F.Brodhun,
and
I.Feussner
(2011).
Oxylipins in fungi.
|
| |
FEBS J,
278,
1047-1063.
|
 |
|
|
|
|
 |
I.N.Van Bogaert,
S.Groeneboer,
K.Saerens,
and
W.Soetaert
(2011).
The role of cytochrome P450 monooxygenases in microbial fatty acid metabolism.
|
| |
FEBS J,
278,
206-221.
|
 |
|
|
|
|
 |
S.Forêt,
F.Seneca,
D.de Jong,
A.Bieller,
G.Hemmrich,
R.Augustin,
D.C.Hayward,
E.E.Ball,
T.C.Bosch,
K.Agata,
M.Hassel,
and
D.J.Miller
(2011).
Phylogenomics reveals an anomalous distribution of USP genes in metazoans.
|
| |
Mol Biol Evol,
28,
153-161.
|
 |
|
|
|
|
 |
X.Wang
(2011).
Structure, function, and engineering of enzymes in isoflavonoid biosynthesis.
|
| |
Funct Integr Genomics,
11,
13-22.
|
 |
|
|
|
|
 |
F.Jernerén,
A.Sesma,
M.Franceschetti,
M.Francheschetti,
M.Hamberg,
and
E.H.Oliw
(2010).
Gene deletion of 7,8-linoleate diol synthase of the rice blast fungus: studies on pathogenicity, stereochemistry, and oxygenation mechanisms.
|
| |
J Biol Chem,
285,
5308-5316.
|
 |
|
|
|
|
 |
L.E.Thornton,
S.G.Rupasinghe,
H.Peng,
M.A.Schuler,
and
M.M.Neff
(2010).
Arabidopsis CYP72C1 is an atypical cytochrome P450 that inactivates brassinosteroids.
|
| |
Plant Mol Biol,
74,
167-181.
|
 |
|
|
|
|
 |
M.Mizutani,
and
D.Ohta
(2010).
Diversification of P450 genes during land plant evolution.
|
| |
Annu Rev Plant Biol,
61,
291-315.
|
 |
|
|
|
|
 |
T.C.Pochapsky,
S.Kazanis,
and
M.Dang
(2010).
Conformational plasticity and structure/function relationships in cytochromes P450.
|
| |
Antioxid Redox Signal,
13,
1273-1296.
|
 |
|
|
|
|
 |
Y.Y.Toporkova,
E.V.Osipova,
L.S.h.Mukhtarova,
Y.V.Gogolev,
and
A.N.Grechkin
(2010).
Alteration of catalysis of CYP74C subfamily enzymes as a result of site-directed mutagenesis.
|
| |
Dokl Biochem Biophys,
435,
287-290.
|
 |
|
|
|
|
 |
A.J.Koo,
and
G.A.Howe
(2009).
The wound hormone jasmonate.
|
| |
Phytochemistry,
70,
1571-1580.
|
 |
|
|
|
|
 |
A.R.Brash
(2009).
Mechanistic aspects of CYP74 allene oxide synthases and related cytochrome P450 enzymes.
|
| |
Phytochemistry,
70,
1522-1531.
|
 |
|
|
|
|
 |
B.Gao,
W.E.Boeglin,
Y.Zheng,
C.Schneider,
and
A.R.Brash
(2009).
Evidence for an ionic intermediate in the transformation of fatty acid hydroperoxide by a catalase-related allene oxide synthase from the Cyanobacterium Acaryochloris marina.
|
| |
J Biol Chem,
284,
22087-22098.
|
 |
|
|
|
|
 |
F.Brodhun,
C.Göbel,
E.Hornung,
and
I.Feussner
(2009).
Identification of PpoA from Aspergillus nidulans as a Fusion Protein of a Fatty Acid Heme Dioxygenase/Peroxidase and a Cytochrome P450.
|
| |
J Biol Chem,
284,
11792-11805.
|
 |
|
|
|
|
 |
G.d'Ippolito,
N.Lamari,
M.Montresor,
G.Romano,
A.Cutignano,
A.Gerecht,
G.Cimino,
and
A.Fontana
(2009).
15S-lipoxygenase metabolism in the marine diatom Pseudo-nitzschia delicatissima.
|
| |
New Phytol,
183,
1064-1071.
|
 |
|
|
|
|
 |
I.R.Chechetkin,
F.K.Mukhitova,
A.S.Blufard,
A.Y.Yarin,
L.L.Antsygina,
and
A.N.Grechkin
(2009).
Unprecedented pathogen-inducible complex oxylipins from flax--linolipins A and B.
|
| |
FEBS J,
276,
4463-4472.
|
 |
|
|
|
|
 |
J.Browse
(2009).
Jasmonate passes muster: a receptor and targets for the defense hormone.
|
| |
Annu Rev Plant Biol,
60,
183-205.
|
 |
|
|
|
|
 |
T.K.Yanai,
and
S.Mori
(2009).
Density functional studies on isomerization of prostaglandin H2 to prostacyclin catalyzed by cytochrome P450.
|
| |
Chemistry,
15,
4464-4473.
|
 |
|
|
|
|
 |
L.J.Marnett
(2008).
Biochemistry: Divergence from the superfamily.
|
| |
Nature,
455,
300-301.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

