 |
PDBsum entry 3bar

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.23
- orotidine-5'-phosphate decarboxylase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
orotidine 5'-phosphate + H+ = UMP + CO2
|
 |
 |
 |
 |
 |
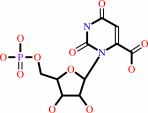 orotidine 5'-phosphate
orotidine 5'-phosphate
|
+
|
H(+)
|
=
|
 UMP
UMP
|
+
|
CO2
Bound ligand (Het Group name = )
corresponds exactly
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Med Chem
51:439-448
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure-activity relationships of C6-uridine derivatives targeting plasmodia orotidine monophosphate decarboxylase.
|
|
A.M.Bello,
E.Poduch,
Y.Liu,
L.Wei,
I.Crandall,
X.Wang,
C.Dyanand,
K.C.Kain,
E.F.Pai,
L.P.Kotra.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Malaria, caused by Plasmodia parasites, has re-emerged as a major problem,
imposing its fatal effects on human health, especially due to multidrug
resistance. In Plasmodia, orotidine 5'-monophosphate decarboxylase (ODCase) is
an essential enzyme for the de novo synthesis of uridine 5'-monophosphate.
Impairing ODCase in these pathogens is a promising strategy to develop novel
classes of therapeutics. Encouraged by our recent discovery that 6-iodo uridine
is a potent inhibitor of P. falciparum, we investigated the structure-activity
relationships of various C6 derivatives of UMP. 6-Cyano, 6-azido, 6-amino,
6-methyl, 6- N-methylamino, and 6- N, N-dimethylamino derivatives of uridine
were evaluated against P. falciparum. The mononucleotides of 6-cyano, 6-azido,
6-amino, and 6-methyl uridine derivatives were studied as inhibitors of
plasmodial ODCase. 6-Azidouridine 5'-monophosphate is a potent covalent
inhibitor of P. falciparum ODCase. 6-Methyluridine exhibited weak antimalarial
activity against P. falciparum 3D7 isolate. 6- N-Methylamino and 6- N,
N-dimethylamino uridine derivatives exhibited moderate antimalarial activities.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
C.Huthmacher,
A.Hoppe,
S.Bulik,
and
H.G.Holzhütter
(2010).
Antimalarial drug targets in Plasmodium falciparum predicted by stage-specific metabolic network analysis.
|
| |
BMC Syst Biol,
4,
120.
|
 |
|
|
|
|
 |
M.A.Phillips,
and
P.K.Rathod
(2010).
Plasmodium dihydroorotate dehydrogenase: a promising target for novel anti-malarial chemotherapy.
|
| |
Infect Disord Drug Targets,
10,
226-239.
|
 |
|
|
|
|
 |
D.Heinrich,
U.Diederichsen,
and
M.G.Rudolph
(2009).
Lys314 is a nucleophile in non-classical reactions of orotidine-5'-monophosphate decarboxylase.
|
| |
Chemistry,
15,
6619-6625.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
J.H.Wong,
U.Sahni,
Y.Li,
X.Chen,
and
J.Gervay-Hague
(2009).
Synthesis of sulfone-based nucleotide isosteres: identification of CMP-sialic acid synthetase inhibitors.
|
| |
Org Biomol Chem,
7,
27-29.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

