 |
PDBsum entry 3gdf

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
3gdf
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of the NADP-dependent mannitol dehydrogenase from cladosporium herbarum.
|
|
Structure:
|
 |
Probable NADP-dependent mannitol dehydrogenase. Chain: a, b, c, d. Synonym: mtdh, mannitol 2-dehydrogenase [nadp+]. Engineered: yes
|
|
Source:
|
 |
Cladosporium herbarum. Organism_taxid: 29918. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.181
|
R-free:
|
0.262
|
|
|
Authors:
|
 |
D.Nuess,P.Goettig,I.Magler,U.Denk,M.Breitenbach,P.B.Schneider, H.Brandstetter,B.Simon-Nobbe
|
|
Key ref:
|
 |
D.Nüss
et al.
(2010).
Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum: Implications for oligomerisation and catalysis.
Biochimie,
92,
985-993.
PubMed id:
|
 |
|
Date:
|
 |
|
24-Feb-09
|
Release date:
|
26-May-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0C0Y5
(MTDH_DAVTA) -
NADP-dependent mannitol dehydrogenase from Davidiella tassiana
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
267 a.a.
267 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.1.1.138
- mannitol 2-dehydrogenase (NADP(+)).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
D-mannitol + NADP+ = D-fructose + NADPH + H+
|
 |
 |
 |
 |
 |
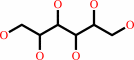 D-mannitol
D-mannitol
|
+
|
 NADP(+)
NADP(+)
|
=
|
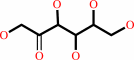 D-fructose
D-fructose
|
+
|
 NADPH
NADPH
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Biochimie
92:985-993
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of the NADP-dependent mannitol dehydrogenase from Cladosporium herbarum: Implications for oligomerisation and catalysis.
|
|
D.Nüss,
P.Goettig,
I.Magler,
U.Denk,
M.Breitenbach,
P.B.Schneider,
H.Brandstetter,
B.Simon-Nobbe.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The ascomycete Cladosporium herbarum is a prominent fungal inducer of Type I
allergy. The only major allergen identified so far is Cla h 8, a NADP-dependent
mannitol dehydrogenase (MtDH). MtDH, a cytoplasmic protein of 28.5kDa, belongs
to the Short chain Dehydrogenases/Reductases (SDR), acting as a NADP-dependent
oxidoreductase. In this study, we found that C. herbarum MtDH can exist as
monomers, dimers and tetramers in solution and, correspondingly, forms tetramers
and higher oligomers in two crystal structures. Additionally, we identified a
unique adaptive binding site for the metal ions Na(+) and Zn(2+) that were
distinguished by an anomalous dispersion experiment. A
Translation-Libration-Screw analysis confirmed the stabilising effect of Zn(2+)
for the tetrameric assembly. Moreover, the zinc containing structure explains
the mode of MtDH multimerisation by metal bridging of the tetramers. The
formation of oligomers and higher multimers of MtDH provides a missing link to
its allergenic properties. Based on the well defined active site region and a
comparative analysis with related structures, we can also clarify the atypical
enzymatic properties of MtDH by two alternative binding modes of the substrate
to the active site.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
V.Kallnik,
C.Schulz,
C.Schultz,
P.Schweiger,
and
U.Deppenmeier
(2011).
Properties of recombinant Strep-tagged and untagged hyperthermophilic D-arabitol dehydrogenase from Thermotoga maritima.
|
| |
Appl Microbiol Biotechnol,
90,
1285-1293.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

