 |
PDBsum entry 3e2p

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Catalytic subunit of m. Jannaschii aspartate transcarbamoylase in an orthorhombic crystal form
|
|
Structure:
|
 |
Aspartate carbamoyltransferase. Chain: a, b, c, d, e, f, i, j, k, l, m, n. Fragment: catalytic subunit. Synonym: aspartate transcarbamylase, atcase. Engineered: yes
|
|
Source:
|
 |
Methanocaldococcus jannaschii. Methanococcus jannaschii. Organism_taxid: 2190. Gene: pyrb, mj1581. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
3.00Å
|
R-factor:
|
0.215
|
R-free:
|
0.269
|
|
|
Authors:
|
 |
J.Vitali,M.J.Colaneri
|
|
Key ref:
|
 |
J.Vitali
and
M.J.Colaneri
(2008).
Structure of the catalytic trimer of Methanococcus jannaschii aspartate transcarbamoylase in an orthorhombic crystal form.
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
776-780.
PubMed id:
|
 |
|
Date:
|
 |
|
06-Aug-08
|
Release date:
|
10-Mar-09
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q58976
(PYRB_METJA) -
Aspartate carbamoyltransferase from Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
306 a.a.
306 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.1.3.2
- aspartate carbamoyltransferase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Pyrimidine Biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
carbamoyl phosphate + L-aspartate = N-carbamoyl-L-aspartate + phosphate + H+
|
 |
 |
 |
 |
 |
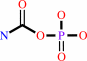 carbamoyl phosphate
carbamoyl phosphate
|
+
|
 L-aspartate
L-aspartate
|
=
|
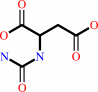 N-carbamoyl-L-aspartate
N-carbamoyl-L-aspartate
|
+
|
 phosphate
phosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Acta Crystallogr Sect F Struct Biol Cryst Commun
64:776-780
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of the catalytic trimer of Methanococcus jannaschii aspartate transcarbamoylase in an orthorhombic crystal form.
|
|
J.Vitali,
M.J.Colaneri.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Crystals of the catalytic subunit of Methanococcus jannaschii aspartate
transcarbamoylase in an orthorhombic crystal form contain four
crystallographically independent trimers which associate in pairs to form stable
staggered complexes that are similar to each other and to a previously
determined monoclinic C2 form. Each subunit has a sulfate in the central
channel. The catalytic subunits in these complexes show flexibility, with the
elbow angles of the monomers differing by up to 7.4 degrees between crystal
forms. Moreover, there is also flexibility in the relative orientation of the
trimers around their threefold axis in the complexes, with a difference of 4
degrees between crystal forms.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

