 |
PDBsum entry 3c3e

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase
|
 |
|
Title:
|
 |
Crystal structure of 2-phospho-(s)-lactate transferase from methanosarcina mazei in complex with fo and gdp. Northeast structural genomics consortium target mar46
|
|
Structure:
|
 |
2-phospho-l-lactate transferase. Chain: a, b, c, d. Synonym: lppg:fo, 2-phospho-(s)-lactate transferase. Engineered: yes
|
|
Source:
|
 |
Methanosarcina mazei go1. Organism_taxid: 192952. Strain: go1 / dsm 3647 / goe1 / jcm 11883 / ocm 88. Atcc: baa-159. Gene: cofd, mm_1874. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
3.00Å
|
R-factor:
|
0.206
|
R-free:
|
0.237
|
|
|
Authors:
|
 |
F.Forouhar,M.Abashidze,H.Xu,L.L.Grochowski,J.Seetharaman,M.Hussain, A.P.Kuzin,Y.Chen,W.Zhou,R.Xiao,T.B.Acton,G.T.Montelione,A.Galinier, R.H.White,L.Tong,Northeast Structural Genomics Consortium (Nesg)
|
Key ref:

|
 |
F.Forouhar
et al.
(2008).
Molecular insights into the biosynthesis of the F420 coenzyme.
J Biol Chem,
283,
11832-11840.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
28-Jan-08
|
Release date:
|
19-Feb-08
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q8PVT6
(COFD_METMA) -
2-phospho-L-lactate transferase from Methanosarcina mazei (strain ATCC BAA-159 / DSM 3647 / Goe1 / Go1 / JCM 11833 / OCM 88)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
303 a.a.
305 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.7.8.28
- 2-phospho-L-lactate transferase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
(2S)-lactyl-2-diphospho-5'-guanosine + 7,8-didemethyl-8-hydroxy-5- deazariboflavin = oxidized coenzyme F420-0 + GMP + H+
|
|
2.
|
enolpyruvoyl-2-diphospho-5'-guanosine + 7,8-didemethyl-8-hydroxy-5- deazariboflavin = dehydro coenzyme F420-0 + GMP + H+
|
|
3.
|
3-[(R)-glyceryl]-diphospho-5'-guanosine + 7,8-didemethyl-8-hydroxy-5- deazariboflavin = oxidized 3PG-factor420-0 + GMP + H+
|
|
 |
 |
 |
 |
 |
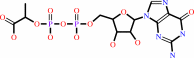 (2S)-lactyl-2-diphospho-5'-guanosine
(2S)-lactyl-2-diphospho-5'-guanosine
|
+
|
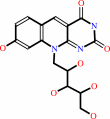 7,8-didemethyl-8-hydroxy-5- deazariboflavin
7,8-didemethyl-8-hydroxy-5- deazariboflavin
|
=
|
oxidized coenzyme F420-0
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
+
|
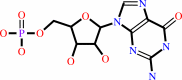 GMP
GMP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
enolpyruvoyl-2-diphospho-5'-guanosine
|
+
|
 7,8-didemethyl-8-hydroxy-5- deazariboflavin
7,8-didemethyl-8-hydroxy-5- deazariboflavin
|
=
|
dehydro coenzyme F420-0
|
+
|
GMP
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
3-[(R)-glyceryl]-diphospho-5'-guanosine
|
+
|
 7,8-didemethyl-8-hydroxy-5- deazariboflavin
7,8-didemethyl-8-hydroxy-5- deazariboflavin
|
=
|
oxidized 3PG-factor420-0
Bound ligand (Het Group name = )
matches with 85.71% similarity
|
+
|
 GMP
GMP
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
283:11832-11840
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Molecular insights into the biosynthesis of the F420 coenzyme.
|
|
F.Forouhar,
M.Abashidze,
H.Xu,
L.L.Grochowski,
J.Seetharaman,
M.Hussain,
A.Kuzin,
Y.Chen,
W.Zhou,
R.Xiao,
T.B.Acton,
G.T.Montelione,
A.Galinier,
R.H.White,
L.Tong.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Coenzyme F(420), a hydride carrier, is found in Archaea and some bacteria and
has crucial roles in methanogenesis, antibiotic biosynthesis, DNA repair, and
activation of antitubercular compounds. CofD, 2-phospho-l-lactate transferase,
catalyzes the last step in the biosynthesis of F(420)-0 (F(420) without
polyglutamate), by transferring the lactyl phosphate moiety of
lactyl(2)diphospho-(5')guanosine to 7,8-didemethyl-8-hydroxy-5-deazariboflavin
ribitol (Fo). CofD is highly conserved among F(420)-producing organisms, and
weak sequence homologs are also found in non-F(420)-producing organisms. This
superfamily does not share any recognizable sequence conservation with other
proteins. Here we report the first crystal structures of CofD, the free enzyme
and two ternary complexes, with Fo and P(i) or with Fo and GDP, from
Methanosarcina mazei. The active site is located at the C-terminal end of a
Rossmann fold core, and three large insertions make significant contributions to
the active site and dimer formation. The observed binding modes of Fo and GDP
can explain known biochemical properties of CofD and are also supported by our
binding assays. The structures provide significant molecular insights into the
biosynthesis of the F(420) coenzyme. Large structural differences in the active
site region of the non-F(420)-producing CofD homologs suggest that they catalyze
a different biochemical reaction.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIGURE 3. Binding mode of GDP. A, final 2F[o] - F[c]
electron density for GDP molecule, contoured at 1  .
Produced with PyMol (41). B, schematic drawing of the
interactions between GDP and CofD. C, overlay of the structure
of the ternary complex with Fo and GDP (in purple) and that with
Fo and P[i] (in yellow). The P[i] is located about 1 Å
from the .
Produced with PyMol (41). B, schematic drawing of the
interactions between GDP and CofD. C, overlay of the structure
of the ternary complex with Fo and GDP (in purple) and that with
Fo and P[i] (in yellow). The P[i] is located about 1 Å
from the  -phosphate of GDP, but
there is a large difference in the conformation of the
glycine-rich loop, indicated with the red star. -phosphate of GDP, but
there is a large difference in the conformation of the
glycine-rich loop, indicated with the red star.
|
 |
Figure 4.
FIGURE 4. Binding mode of Fo. A, final 2F[o] - F[c]
electron density for Fo, contoured at 1  . Produced with PyMol
(41). B, schematic drawing of the interactions between Fo and
CofD. . Produced with PyMol
(41). B, schematic drawing of the interactions between Fo and
CofD.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2008,
283,
11832-11840)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
E.Foulquier,
F.Pompeo,
A.Bernadac,
L.Espinosa,
and
A.Galinier
(2011).
The YvcK protein is required for morphogenesis via localization of PBP1 under gluconeogenic growth conditions in Bacillus subtilis.
|
| |
Mol Microbiol,
80,
309-318.
|
 |
|
|
|
|
 |
J.Luciano,
E.Foulquier,
J.R.Fantino,
A.Galinier,
and
F.Pompeo
(2009).
Characterization of YvcJ, a conserved P-loop-containing protein, and its implication in competence in Bacillus subtilis.
|
| |
J Bacteriol,
191,
1556-1564.
|
 |
|
|
|
|
 |
M.M.Otte,
and
J.C.Escalante-Semerena
(2009).
Biochemical characterization of the GTP:adenosylcobinamide-phosphate guanylyltransferase (CobY) enzyme of the hyperthermophilic archaeon Methanocaldococcus jannaschii.
|
| |
Biochemistry,
48,
5882-5889.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

