 |
PDBsum entry 3a6d

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Creatininase complexed with 1-methylguanidine
|
|
Structure:
|
 |
Creatinine amidohydrolase. Chain: a, b, c, d, e, f. Synonym: creatininase. Engineered: yes
|
|
Source:
|
 |
Pseudomonas putida. Organism_taxid: 303. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
1.90Å
|
R-factor:
|
0.192
|
R-free:
|
0.207
|
|
|
Authors:
|
 |
Y.Nakajima,K.Yamashita,K.Ito,T.Yoshimoto
|
|
Key ref:
|
 |
K.Yamashita
et al.
(2010).
Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase.
J Mol Biol,
396,
1081-1096.
PubMed id:
|
 |
|
Date:
|
 |
|
31-Aug-09
|
Release date:
|
09-Feb-10
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P83772
(CRNA_PSEPU) -
Creatinine amidohydrolase from Pseudomonas putida
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
260 a.a.
257 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.5.2.10
- creatininase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
creatinine + H2O = creatine
|
 |
 |
 |
 |
 |
creatinine
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
H2O
|
=
|
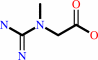 creatine
creatine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
J Mol Biol
396:1081-1096
(2010)
|
|
PubMed id:
|
|
|
|

|
| |
|
Substitution of Glu122 by glutamine revealed the function of the second water molecule as a proton donor in the binuclear metal enzyme creatininase.
|
|
K.Yamashita,
Y.Nakajima,
H.Matsushita,
Y.Nishiya,
R.Yamazawa,
Y.F.Wu,
F.Matsubara,
H.Oyama,
K.Ito,
T.Yoshimoto.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Creatininase is a binuclear zinc enzyme and catalyzes the reversible conversion
of creatinine to creatine. It exhibits an open-closed conformational change upon
substrate binding, and the differences in the conformations of Tyr121, Trp154,
and the loop region containing Trp174 were evident in the enzyme-creatine
complex when compared to those in the ligand-free enzyme. We have determined the
crystal structure of the enzyme complexed with a 1-methylguanidine. All subunits
in the complex existed as the closed form, and the binding mode of creatinine
was estimated. Site-directed mutagenesis revealed that the hydrophobic residues
that show conformational change upon substrate binding are important for the
enzyme activity. We propose a catalytic mechanism of creatininase in which two
water molecules have significant roles. The first molecule is a hydroxide ion
(Wat1) that is bound as a bridge between the two metal ions and attacks the
carbonyl carbon of the substrate. The second molecule is a water molecule (Wat2)
that is bound to the carboxyl group of Glu122 and functions as a proton donor in
catalysis. The activity of the E122Q mutant was very low and it was only
partially restored by the addition of ZnCl(2) or MnCl(2). In the E122Q mutant,
k(cat) is drastically decreased, indicating that Glu122 is important for
catalysis. X-ray crystallographic study and the atomic absorption spectrometry
analysis of the E122Q mutant-substrate complex revealed that the drastic
decrease of the activity of the E122Q was caused by not only the loss of one Zn
ion at the Metal1 site but also a critical function of Glu122, which most likely
exists for a proton transfer step through Wat2.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

