 |
PDBsum entry 2vq7

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2vq7
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.13.148
- trimethylamine monooxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
trimethylamine + NADPH + O2 = trimethylamine N-oxide + NADP+ + H2O
|
 |
 |
 |
 |
 |
trimethylamine
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADPH
NADPH
|
+
|
 O2
O2
|
=
|
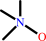 trimethylamine N-oxide
trimethylamine N-oxide
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Flavoprotein
|
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
105:6572-6577
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Revealing the moonlighting role of NADP in the structure of a flavin-containing monooxygenase.
|
|
A.Alfieri,
E.Malito,
R.Orru,
M.W.Fraaije,
A.Mattevi.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Flavin-containing monooxygenases (FMOs) are, after cytochromes P450, the most
important monooxygenase system in humans and are involved in xenobiotics
metabolism and variability in drug response. The x-ray structure of a soluble
prokaryotic FMO from Methylophaga sp. strain SK1 has been solved at 2.6-A
resolution and is now the protein of known structure with the highest sequence
similarity to human FMOs. The structure possesses a two-domain architecture,
with both FAD and NADP(+) well defined by the electron density maps. Biochemical
analysis shows that the prokaryotic enzyme shares many functional properties
with mammalian FMOs, including substrate specificity and the ability to
stabilize the hydroperoxyflavin intermediate that is crucial in substrate
oxygenation. On the basis of their location in the structure, the nicotinamide
ring and the adjacent ribose of NADP(+) turn out to be an integral part of the
catalytic site being actively engaged in the stabilization of the oxygenating
intermediate. This feature suggests that NADP(H) has a moonlighting role, in
that it adopts two binding modes that allow it to function in both flavin
reduction and oxygen reactivity modulation, respectively. We hypothesize that a
relative domain rotation is needed to bring NADP(H) to these distinct positions
inside the active site. Localization of mutations in human FMO3 that are known
to cause trimethylaminuria (fish-odor syndrome) in the elucidated FMO structure
provides a structural explanation for their biological effects.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Overall crystal structure of mFMO. (A) Ribbon diagram of the
monomer. FAD-binding domain (residues 8–169 and 281–450) is
orange and NADP-binding domain (residues 170–280) is green.
FAD is shown as yellow sticks and NADP^+ as blue sticks. The
positions of the long interdomain loop (residues 44–80), the
hinge connecting the two domains, and the polypeptide stretch
corresponding to residues 407–415 are outlined. mFMO residues
corresponding to TMAU-causing mutations (17) and polymorphisms
in hFMO3 (in parentheses) are in red and blue sticks,
respectively. The position of a long insert in hFMO3 (residues
238–299; Fig. 1B) is also indicated. It is expected to occupy
a surface-exposed position away from the active site. (B) Ribbon
representation of the mFMO dimer. One monomer is shown in the
same orientation and color as Fig. 3A; the other one is colored
gray, with the NADP-binding domain in darker gray.
|
 |
Figure 5.
The role of NADP^+ in the stabilization of
C4a-hydroperoxyflavin intermediate. (A) Modeling experiment in
which the hypothetical structure of C4a-hydroperoxyflavin was
superimposed to the flavin in mFMO structure. The color code is
the same as in Fig. 4B. Hypothetical hydrogen bonds involving
the hydroperoxyflavin atoms are shown as blue dashed lines. The
accommodation of the additional oxygen atoms of the C4a-adduct
would require a shift of ≈1.5 Å of Asn-78 side chain
(whose conformation in the native structure is shown as thin
black stick). (B) Comparison of the NADP^+-binding mode in S.
pombe (Protein Data Bank ID code 2gv8) and Methylophaga FMOs.
The picture was obtained by superimposing the Cα atoms of the
two proteins and shows the FAD (yellow) and NADP^+ (blue)
molecules of mFMO together with FAD and NADP^+ of the S. pombe
enzyme (red). The N5 and C4a atoms of the flavin and C4 and C2
atoms of NADP^+ are labeled.
|
 |
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
A.Rioz-Martínez,
M.Kopacz,
G.de Gonzalo,
D.E.Torres Pazmiño,
V.Gotor,
and
M.W.Fraaije
(2011).
Exploring the biocatalytic scope of a bacterial flavin-containing monooxygenase.
|
| |
Org Biomol Chem,
9,
1337-1341.
|
 |
|
|
|
|
 |
H.J.Cho,
H.Y.Cho,
K.J.Kim,
M.H.Kim,
S.W.Kim,
and
B.S.Kang
(2011).
Structural and functional analysis of bacterial flavin-containing monooxygenase reveals its ping-pong-type reaction mechanism.
|
| |
J Struct Biol,
175,
39-48.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
H.M.Dudek,
D.E.Torres Pazmiño,
C.Rodríguez,
G.de Gonzalo,
V.Gotor,
and
M.W.Fraaije
(2010).
Investigating the coenzyme specificity of phenylacetone monooxygenase from Thermobifida fusca.
|
| |
Appl Microbiol Biotechnol,
88,
1135-1143.
|
 |
|
|
|
|
 |
S.C.Li,
C.Y.Lin,
T.F.Kuo,
Y.H.Lin,
C.C.Chen,
W.N.Lin,
and
W.P.Chan
(2010).
Chicken model of steroid-induced bone marrow adipogenesis using proteome analysis: a preliminary study.
|
| |
Proteome Sci,
8,
47.
|
 |
|
|
|
|
 |
S.Chakraborty,
M.Ortiz-Maldonado,
B.Entsch,
and
D.P.Ballou
(2010).
Studies on the mechanism of p-hydroxyphenylacetate 3-hydroxylase from Pseudomonas aeruginosa: a system composed of a small flavin reductase and a large flavin-dependent oxygenase.
|
| |
Biochemistry,
49,
372-385.
|
 |
|
|
|
|
 |
S.Sehlmeyer,
L.Wang,
D.Langel,
D.G.Heckel,
H.Mohagheghi,
G.Petschenka,
and
D.Ober
(2010).
Flavin-dependent monooxygenases as a detoxification mechanism in insects: new insights from the arctiids (lepidoptera).
|
| |
PLoS One,
5,
e10435.
|
 |
|
|
|
|
 |
F.Forneris,
R.Orru,
D.Bonivento,
L.R.Chiarelli,
and
A.Mattevi
(2009).
ThermoFAD, a Thermofluor-adapted flavin ad hoc detection system for protein folding and ligand binding.
|
| |
FEBS J,
276,
2833-2840.
|
 |
|
|
|
|
 |
M.S.Motika,
J.Zhang,
X.Zheng,
K.Riedler,
and
J.R.Cashman
(2009).
Novel variants of the human flavin-containing monooxygenase 3 (FMO3) gene associated with trimethylaminuria.
|
| |
Mol Genet Metab,
97,
128-135.
|
 |
|
|
|
|
 |
S.K.Krueger,
M.C.Henderson,
L.K.Siddens,
J.E.VanDyke,
A.D.Benninghoff,
P.A.Karplus,
B.Furnes,
D.Schlenk,
and
D.E.Williams
(2009).
Characterization of sulfoxygenation and structural implications of human flavin-containing monooxygenase isoform 2 (FMO2.1) variants S195L and N413K.
|
| |
Drug Metab Dispos,
37,
1785-1791.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

