 |
PDBsum entry 2v0c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Ligase
|
 |
|
Title:
|
 |
Leucyl-tRNA synthetase from thermus thermophilus complexed with a sulphamoyl analogue of leucyl-adenylate in the synthetic site and an adduct of amp with 5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (an2690) in the editing site
|
|
Structure:
|
 |
Aminoacyl-tRNA synthetase. Chain: a. Synonym: leucyl-tRNA synthetase. Engineered: yes
|
|
Source:
|
 |
Thermus thermophilus. Organism_taxid: 274. Strain: hb-27. Expressed in: escherichia coli. Expression_system_taxid: 469008.
|
|
Resolution:
|
 |
|
1.85Å
|
R-factor:
|
0.181
|
R-free:
|
0.202
|
|
|
Authors:
|
 |
F.Rock,W.Mao,A.Yaremchuk,M.Tukalo,T.Crepin,H.Zhou,Y.Zhang, V.Hernandez,T.Akama,S.Baker,J.Plattner,L.Shapiro,S.A.Martinis, S.J.Benkovic,S.Cusack,M.R.K.Alley
|
Key ref:

|
 |
F.L.Rock
et al.
(2007).
An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site.
Science,
316,
1759-1761.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
14-May-07
|
Release date:
|
03-Jul-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q7SIE4
(Q7SIE4_THETH) -
Leucine--tRNA ligase from Thermus thermophilus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
878 a.a.
810 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.6.1.1.4
- leucine--tRNA ligase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
tRNA(Leu) + L-leucine + ATP = L-leucyl-tRNA(Leu) + AMP + diphosphate
|
 |
 |
 |
 |
 |
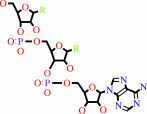 tRNA(Leu)
tRNA(Leu)
|
+
|
L-leucine
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
 ATP
ATP
|
=
|
L-leucyl-tRNA(Leu)
Bound ligand (Het Group name = )
matches with 69.70% similarity
|
+
|
 AMP
AMP
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Science
316:1759-1761
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
An antifungal agent inhibits an aminoacyl-tRNA synthetase by trapping tRNA in the editing site.
|
|
F.L.Rock,
W.Mao,
A.Yaremchuk,
M.Tukalo,
T.Crépin,
H.Zhou,
Y.K.Zhang,
V.Hernandez,
T.Akama,
S.J.Baker,
J.J.Plattner,
L.Shapiro,
S.A.Martinis,
S.J.Benkovic,
S.Cusack,
M.R.Alley.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aminoacyl-transfer RNA (tRNA) synthetases, which catalyze the attachment of the
correct amino acid to its corresponding tRNA during translation of the genetic
code, are proven antimicrobial drug targets. We show that the broad-spectrum
antifungal 5-fluoro-1,3-dihydro-1-hydroxy-2,1-benzoxaborole (AN2690), in
development for the treatment of onychomycosis, inhibits yeast cytoplasmic
leucyl-tRNA synthetase by formation of a stable tRNA(Leu)-AN2690 adduct in the
editing site of the enzyme. Adduct formation is mediated through the boron atom
of AN2690 and the 2'- and 3'-oxygen atoms of tRNA's3'-terminal adenosine. The
trapping of enzyme-bound tRNA(Leu) in the editing site prevents catalytic
turnover, thus inhibiting synthesis of leucyl-tRNA(Leu) and consequentially
blocking protein synthesis. This result establishes the editing site as a bona
fide target for aminoacyl-tRNA synthetase inhibitors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Fig. 3. AN2690 forms an adduct with the terminal adenosine
(A76) of tRNA^Leu in the editing active site of LeuRS. (A)
Overall structure of the complex of T. thermophilus LeuRS with
tRNA^Leu and AN2690, showing the adenosine-AN2690 adduct
(ball-and-stick model, ringed in red) in the editing site and
leucine (space-filling model) in the synthetic site. The editing
domain is cyan; the catalytic domain, yellow; Zn-1 domain,
purple; the leucyl-specific insertion domain, black; the
anticodon-binding domain, red; the C-terminal domain, gold; zinc
atoms, gray spheres; and tRNA, blue tube. (B) Unbiased
difference map (1.85 Å resolution) for the AMP-AN2690
adduct in the editing site. (C) Diagram showing water molecules
(dark blue spheres) and hydrogen bonds (green dotted lines)
between editing site residues of LeuRS and the AMP-AN2690 adduct
(orange). Amino acid residues that are mutated in the S.
cerevisiae AN2690-resistant mutants are labeled and colored in
purple (table S2). The atoms are colored accordingly: boron,
mauve; fluorine, green; oxygen, red; nitrogen, light blue;
carbon, yellow; and phosphate, purple. (D) Superposition of
bound posttransfer editing substrate analog (Nva2AA, brown) (9)
and the AMP-AN2690 adduct (orange) obtained after superposing
the C  positions of the
editing domain of each complex. positions of the
editing domain of each complex.
|
 |
Figure 4.
Fig. 4. Boron and the oxaborole ring are required for
inhibition of aminoacylation. All reactions were performed in
triplicate, and the mean values were used to determine a median
inhibitory concentration (IC[50]) with Prism 4 (17).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the AAAs:
Science
(2007,
316,
1759-1761)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
H.Zhou,
L.Sun,
X.L.Yang,
and
P.Schimmel
(2013).
ATP-directed capture of bioactive herbal-based medicine on human tRNA synthetase.
|
| |
Nature,
494,
121-124.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
A.Palencia,
T.Crépin,
M.T.Vu,
T.L.Lincecum,
S.A.Martinis,
and
S.Cusack
(2012).
Structural dynamics of the aminoacylation and proofreading functional cycle of bacterial leucyl-tRNA synthetase.
|
| |
Nat Struct Mol Biol,
19,
677-684.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
L.Li,
L.Yu,
and
Q.Huang
(2011).
Molecular trigger for pre-transfer editing pathway in Valyl-tRNA synthetase: A molecular dynamics simulation study.
|
| |
J Mol Model,
17,
555-564.
|
 |
|
|
|
|
 |
X.Chen,
J.J.Ma,
M.Tan,
P.Yao,
Q.H.Hu,
G.Eriani,
and
E.D.Wang
(2011).
Modular pathways for editing non-cognate amino acids by human cytoplasmic leucyl-tRNA synthetase.
|
| |
Nucleic Acids Res,
39,
235-247.
|
 |
|
|
|
|
 |
Y.Xia,
K.Cao,
Y.Zhou,
M.R.Alley,
F.Rock,
M.Mohan,
M.Meewan,
S.J.Baker,
S.Lux,
C.Z.Ding,
G.Jia,
M.Kully,
and
J.J.Plattner
(2011).
Synthesis and SAR of novel benzoxaboroles as a new class of β-lactamase inhibitors.
|
| |
Bioorg Med Chem Lett,
21,
2533-2536.
|
 |
|
|
|
|
 |
A.M.Smith,
R.Ammar,
C.Nislow,
and
G.Giaever
(2010).
A survey of yeast genomic assays for drug and target discovery.
|
| |
Pharmacol Ther,
127,
156-164.
|
 |
|
|
|
|
 |
B.Nare,
S.Wring,
C.Bacchi,
B.Beaudet,
T.Bowling,
R.Brun,
D.Chen,
C.Ding,
Y.Freund,
E.Gaukel,
A.Hussain,
K.Jarnagin,
M.Jenks,
M.Kaiser,
L.Mercer,
E.Mejia,
A.Noe,
M.Orr,
R.Parham,
J.Plattner,
R.Randolph,
D.Rattendi,
C.Rewerts,
J.Sligar,
N.Yarlett,
R.Don,
and
R.Jacobs
(2010).
Discovery of novel orally bioavailable oxaborole 6-carboxamides that demonstrate cure in a murine model of late-stage central nervous system african trypanosomiasis.
|
| |
Antimicrob Agents Chemother,
54,
4379-4388.
|
 |
|
|
|
|
 |
J.M.Becker,
S.J.Kauffman,
M.Hauser,
L.Huang,
M.Lin,
S.Sillaots,
B.Jiang,
D.Xu,
and
T.Roemer
(2010).
Pathway analysis of Candida albicans survival and virulence determinants in a murine infection model.
|
| |
Proc Natl Acad Sci U S A,
107,
22044-22049.
|
 |
|
|
|
|
 |
K.Knott,
J.Fishovitz,
S.B.Thorpe,
I.Lee,
and
W.L.Santos
(2010).
N-Terminal peptidic boronic acids selectively inhibit human ClpXP.
|
| |
Org Biomol Chem,
8,
3451-3456.
|
 |
|
|
|
|
 |
M.N.Gwynn,
A.Portnoy,
S.F.Rittenhouse,
and
D.J.Payne
(2010).
Challenges of antibacterial discovery revisited.
|
| |
Ann N Y Acad Sci,
1213,
5.
|
 |
|
|
|
|
 |
M.Tan,
B.Zhu,
X.L.Zhou,
R.He,
X.Chen,
G.Eriani,
and
E.D.Wang
(2010).
tRNA-dependent pre-transfer editing by prokaryotic leucyl-tRNA synthetase.
|
| |
J Biol Chem,
285,
3235-3244.
|
 |
|
|
|
|
 |
M.Zhou,
X.Dong,
N.Shen,
C.Zhong,
and
J.Ding
(2010).
Crystal structures of Saccharomyces cerevisiae tryptophanyl-tRNA synthetase: new insights into the mechanism of tryptophan activation and implications for anti-fungal drug design.
|
| |
Nucleic Acids Res,
38,
3399-3413.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.A.Martinis,
and
M.T.Boniecki
(2010).
The balance between pre- and post-transfer editing in tRNA synthetases.
|
| |
FEBS Lett,
584,
455-459.
|
 |
|
|
|
|
 |
X.L.Zhou,
M.Tan,
M.Wang,
X.Chen,
and
E.D.Wang
(2010).
Post-transfer editing by a eukaryotic leucyl-tRNA synthetase resistant to the broad-spectrum drug AN2690.
|
| |
Biochem J,
430,
325-333.
|
 |
|
|
|
|
 |
A.P.Mascarenhas,
and
S.A.Martinis
(2009).
A glycine hinge for tRNA-dependent translocation of editing substrates to prevent errors by leucyl-tRNA synthetase.
|
| |
FEBS Lett,
583,
3443-3447.
|
 |
|
|
|
|
 |
J.Ling,
N.Reynolds,
and
M.Ibba
(2009).
Aminoacyl-tRNA synthesis and translational quality control.
|
| |
Annu Rev Microbiol,
63,
61-78.
|
 |
|
|
|
|
 |
J.W.Kozarich
(2009).
The biochemistry of disease: desperately seeking syzygy.
|
| |
Annu Rev Biochem,
78,
55-63.
|
 |
|
|
|
|
 |
K.M.Weimer,
B.L.Shane,
M.Brunetto,
S.Bhattacharyya,
and
S.Hati
(2009).
Evolutionary basis for the coupled-domain motions in Thermus thermophilus leucyl-tRNA synthetase.
|
| |
J Biol Chem,
284,
10088-10099.
|
 |
|
|
|
|
 |
R.A.Hellmann,
and
S.A.Martinis
(2009).
Defects in transient tRNA translocation bypass tRNA synthetase quality control mechanisms.
|
| |
J Biol Chem,
284,
11478-11484.
|
 |
|
|
|
|
 |
T.K.Bhatt,
C.Kapil,
S.Khan,
M.A.Jairajpuri,
V.Sharma,
D.Santoni,
F.Silvestrini,
E.Pizzi,
and
A.Sharma
(2009).
A genomic glimpse of aminoacyl-tRNA synthetases in malaria parasite Plasmodium falciparum.
|
| |
BMC Genomics,
10,
644.
|
 |
|
|
|
|
 |
Y.L.Pang,
and
S.A.Martinis
(2009).
A paradigm shift for the amino acid editing mechanism of human cytoplasmic leucyl-tRNA synthetase.
|
| |
Biochemistry,
48,
8958-8964.
|
 |
|
|
|
|
 |
Y.Zeng,
H.Roy,
P.B.Patil,
M.Ibba,
and
S.Chen
(2009).
Characterization of two seryl-tRNA synthetases in albomycin-producing Streptomyces sp. strain ATCC 700974.
|
| |
Antimicrob Agents Chemother,
53,
4619-4627.
|
 |
|
|
|
|
 |
A.P.Mascarenhas,
and
S.A.Martinis
(2008).
Functional segregation of a predicted "hinge" site within the beta-strand linkers of Escherichia coli leucyl-tRNA synthetase.
|
| |
Biochemistry,
47,
4808-4816.
|
 |
|
|
|
|
 |
C.S.Francklyn
(2008).
DNA polymerases and aminoacyl-tRNA synthetases: shared mechanisms for ensuring the fidelity of gene expression.
|
| |
Biochemistry,
47,
11695-11703.
|
 |
|
|
|
|
 |
K.E.Splan,
K.Musier-Forsyth,
M.T.Boniecki,
and
S.A.Martinis
(2008).
In vitro assays for the determination of aminoacyl-tRNA synthetase editing activity.
|
| |
Methods,
44,
119-128.
|
 |
|
|
|
|
 |
M.T.Boniecki,
M.T.Vu,
A.K.Betha,
and
S.A.Martinis
(2008).
CP1-dependent partitioning of pretransfer and posttransfer editing in leucyl-tRNA synthetase.
|
| |
Proc Natl Acad Sci U S A,
105,
19223-19228.
|
 |
|
|
|
|
 |
S.G.Park,
P.Schimmel,
and
S.Kim
(2008).
Aminoacyl tRNA synthetases and their connections to disease.
|
| |
Proc Natl Acad Sci U S A,
105,
11043-11049.
|
 |
|
|
|
|
 |
J.G.Robertson
(2007).
Enzymes as a special class of therapeutic target: clinical drugs and modes of action.
|
| |
Curr Opin Struct Biol,
17,
674-679.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

