 |
PDBsum entry 2phh

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2phh
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.14.13.2
- 4-hydroxybenzoate 3-monooxygenase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Benzoate Metabolism
|
 |
 |
 |
 |
 |
Reaction:
|
 |
4-hydroxybenzoate + NADPH + O2 + H+ = 3,4-dihydroxybenzoate + NADP+ + H2O
|
 |
 |
 |
 |
 |
4-hydroxybenzoate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
 NADPH
NADPH
|
+
|
O2
Bound ligand (Het Group name = )
matches with 71.43% similarity
|
+
|
H(+)
|
=
|
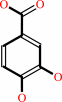 3,4-dihydroxybenzoate
3,4-dihydroxybenzoate
|
+
|
 NADP(+)
NADP(+)
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
FAD
|
 |
 |
 |
 |
 |
 FAD
FAD
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Biochemistry
28:7199-7205
(1989)
|
|
PubMed id:
|
|
|
|

|
| |
|
The coenzyme analogue adenosine 5-diphosphoribose displaces FAD in the active site of p-hydroxybenzoate hydroxylase. An x-ray crystallographic investigation.
|
|
J.M.van der Laan,
H.A.Schreuder,
M.B.Swarte,
R.K.Wierenga,
K.H.Kalk,
W.G.Hol,
J.Drenth.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
p-Hydroxybenzoate hydroxylase (PHBH) is an NADPH-dependent enzyme. To locate the
NADPH binding site, the enzyme was crystallized under anaerobic conditions in
the presence of the substrate p-hydroxybenzoate, the coenzyme analogue adenosine
5-diphosphoribose (ADPR), and sodium dithionite. This yielded colorless crystals
that were suitable for X-ray analysis. Diffraction data were collected up to
2.7-A resolution. A difference Fourier between data from these colorless
crystals and data from yellow crystals of the enzyme-substrate complex showed
that in the colorless crystals the flavin ring was absent. The adenosine
5'-diphosphate moiety, which is the common part between FAD and ADPR, was still
present. After restrained least-squares refinement of the enzyme-substrate
complex with the riboflavin omitted from the model, additional electron density
appeared near the pyrophosphate, which indicated the presence of an ADPR
molecule in the FAD binding site of PHBH. The complete ADPR molecule was fitted
to the electron density, and subsequent least-squares refinement resulted in a
final R factor of 16.8%. Replacement of bound FAD by ADPR was confirmed by
equilibrium dialysis, where it was shown that ADPR can effectively remove FAD
from the enzyme under mild conditions in 0.1 M potassium phosphate buffer, pH
8.0. The empty pocket left by the flavin ring is filled by solvent, leaving the
architecture of the active site and the binding of the substrate largely
unaffected.

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
M.H.Eppink,
H.A.Schreuder,
and
W.J.van Berkel
(1998).
Interdomain binding of NADPH in p-hydroxybenzoate hydroxylase as suggested by kinetic, crystallographic and modeling studies of histidine 162 and arginine 269 variants.
|
| |
J Biol Chem,
273,
21031-21039.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
M.H.Eppink,
H.A.Schreuder,
and
W.J.Van Berkel
(1997).
Identification of a novel conserved sequence motif in flavoprotein hydroxylases with a putative dual function in FAD/NAD(P)H binding.
|
| |
Protein Sci,
6,
2454-2458.
|
 |
|
|
|
|
 |
B.Seibold,
M.Matthes,
M.H.Eppink,
F.Lingens,
W.J.Van Berkel,
and
R.Müller
(1996).
4-Hydroxybenzoate hydroxylase from Pseudomonas sp. CBS3. Purification, characterization, gene cloning, sequence analysis and assignment of structural features determining the coenzyme specificity.
|
| |
Eur J Biochem,
239,
469-478.
|
 |
|
|
|
|
 |
M.H.Eppink,
H.A.Schreuder,
and
W.J.Van Berkel
(1995).
Structure and function of mutant Arg44Lys of 4-hydroxybenzoate hydroxylase implications for NADPH binding.
|
| |
Eur J Biochem,
231,
157-165.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
B.Averhoff,
L.Gregg-Jolly,
D.Elsemore,
and
L.N.Ornston
(1992).
Genetic analysis of supraoperonic clustering by use of natural transformation in Acinetobacter calcoaceticus.
|
| |
J Bacteriol,
174,
200-204.
|
 |
|
|
|
|
 |
H.A.Schreuder,
J.M.van der Laan,
M.B.Swarte,
K.H.Kalk,
W.G.Hol,
and
J.Drenth
(1992).
Crystal structure of the reduced form of p-hydroxybenzoate hydroxylase refined at 2.3 A resolution.
|
| |
Proteins,
14,
178-190.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

