 |
PDBsum entry 2pc5

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.6.1.23
- dUTP diphosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
dUTP + H2O = dUMP + diphosphate + H+
|
 |
 |
 |
 |
 |
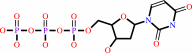 dUTP
dUTP
|
+
|
H2O
|
=
|
 dUMP
dUMP
|
+
|
 diphosphate
diphosphate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
|
Acta Crystallogr Sect F Struct Biol Cryst Commun
63:409-411
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Purification, crystallization and preliminary crystallographic analysis of deoxyuridine triphosphate nucleotidohydrolase from Arabidopsis thaliana.
|
|
M.Bajaj,
H.Moriyama.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The deoxyuridine triphosphate nucleotidohydrolase gene from Arabidopsis thaliana
was expressed and the gene product was purified. Crystallization was performed
by the hanging-drop vapour-diffusion method at 298 K using 2 M ammonium sulfate
as the precipitant. X-ray diffraction data were collected to 2.2 A resolution
using Cu K alpha radiation. The crystal belongs to the orthorhombic space group
P2(1)2(1)2(1), with unit-cell parameters a = 69.90, b = 70.86 A, c = 75.55 A.
Assuming the presence of a trimer in the asymmetric unit, the solvent content
was 30%, with a V(M) of 1.8 A3 Da(-1).

|

|
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Homma,
and
H.Moriyama
(2009).
Crystallization and crystal-packing studies of Chlorella virus deoxyuridine triphosphatase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
65,
1030-1034.
|
 |
|
PDB codes:
|
 |
|
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

