 |
PDBsum entry 2pc0

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.3.4.23.16
- HIV-1 retropepsin.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
Specific for a P1 residue that is hydrophobic, and P1' variable, but often Pro.
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
DOI no:
|
Acta Crystallogr D Biol Crystallogr
63:866-875
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Conformational flexibility in the flap domains of ligand-free HIV protease.
|
|
H.Heaslet,
R.Rosenfeld,
M.Giffin,
Y.C.Lin,
K.Tam,
B.E.Torbett,
J.H.Elder,
D.E.McRee,
C.D.Stout.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of wild-type HIV protease (HIV PR) in the absence of
substrate or inhibitor in two related crystal forms at 1.4 and 2.15 A resolution
are reported. In one crystal form HIV PR adopts an 'open' conformation with a
7.7 A separation between the tips of the flaps in the homodimer. In the other
crystal form the tips of the flaps are 'curled' towards the 80s loop, forming
contacts across the local twofold axis. The 2.3 A resolution crystal structure
of a sixfold mutant of HIV PR in the absence of substrate or inhibitor is also
reported. The mutant HIV PR, which evolved in response to treatment with the
potent inhibitor TL-3, contains six point mutations relative to the wild-type
enzyme (L24I, M46I, F53L, L63P, V77I, V82A). In this structure the flaps also
adopt a 'curled' conformation, but are separated and not in contact. Comparison
of the apo structures to those with TL-3 bound demonstrates the extent of
conformational change induced by inhibitor binding, which includes
reorganization of the packing between twofold-related flaps. Further comparison
with six other apo HIV PR structures reveals that the 'open' and 'curled'
conformations define two distinct families in HIV PR. These conformational
states include hinge motion of residues at either end of the flaps, opening and
closing the entire beta-loop, and translational motion of the flap normal to the
dimer twofold axis and relative to the 80s loop. The alternate conformations
also entail changes in the beta-turn at the tip of the flap. These observations
provide insight into the plasticity of the flap domains, the nature of their
motions and their critical role in binding substrates and inhibitors.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3 Flap conformational changes induced by binding of TL-3
to wild-type HIV PR. In the absence of substrate or inhibitor,
the flap regions of wild-type HIV PR adopt two distinct
conformations: `open' (rose) and `curled' (green). While the
flap regions lack interactions in the `open' conformation, a
stabilizing packing interaction is formed between the side
chains of Phe53 and Ile50' of twofold-related monomers in the
`curled' conformation. Upon binding of TL-3 (cyan), the flaps
shift toward the active site and their interactions are
reconfigured: the side chain of Phe53 now interacts with the P4
benzyl group of TL-3 (light blue) and Ile50 contacts Ile54'.
TL-3 also induces a shift in the 80s loop, which moves towards
the inhibitor such that Pro81' and Val82' contact the P1 benzyl
group.
|
 |
Figure 4.
Figure 4 Expanded `curled' flap conformation in the 2.3 Å
structure of apo 6× HIV PR: view of the flaps, 80s loop
and active site of the unliganded 6× HIV PR showing the
expanded `curled' configuration of the flaps. As in the `curled'
conformation of wild-type HIV PR, the flaps of 6× HIV PR
adopt the `top-to-top' orientation, but are 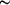 5
Å farther apart and lack stabilizing interactions. The
flap residues Gly48 and Gly49 are disordered, along with the
side chain of Ile50. The further expansion shifts the flaps
toward the 80s loop, allowing a packing contact between Ile50
and Pro81 in the same subunit. The active site is devoid of
ordered water molecules or metal ions. 5
Å farther apart and lack stabilizing interactions. The
flap residues Gly48 and Gly49 are disordered, along with the
side chain of Ile50. The further expansion shifts the flaps
toward the 80s loop, allowing a packing contact between Ile50
and Pro81 in the same subunit. The active site is devoid of
ordered water molecules or metal ions.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the IUCr:
Acta Crystallogr D Biol Crystallogr
(2007,
63,
866-875)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
S.Karthik,
and
S.Senapati
(2011).
Dynamic flaps in HIV-1 protease adopt unique ordering at different stages in the catalytic cycle.
|
| |
Proteins,
79,
1830-1840.
|
 |
|
|
|
|
 |
J.A.Cuesta-Seijo,
C.Neale,
M.A.Khan,
J.Moktar,
C.D.Tran,
R.E.Bishop,
R.Pomès,
and
G.G.Privé
(2010).
PagP crystallized from SDS/cosolvent reveals the route for phospholipid access to the hydrocarbon ruler.
|
| |
Structure,
18,
1210-1219.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
Y.C.Lin,
B.E.Torbett,
and
J.H.Elder
(2010).
Generation of infectious feline immunodeficiency virus (FIV) encoding FIV/human immunodeficiency virus chimeric protease.
|
| |
J Virol,
84,
6799-6809.
|
 |
|
|
|
|
 |
A.J.Kandathil,
A.P.Joseph,
R.Kannangai,
N.Srinivasan,
O.C.Abraham,
S.A.Pulimood,
and
G.Sridharan
(2009).
Structural basis of drug resistance by genetic variants of HIV type 1 clade c protease from India.
|
| |
AIDS Res Hum Retroviruses,
25,
511-519.
|
 |
|
|
|
|
 |
J.L.Kear,
M.E.Blackburn,
A.M.Veloro,
B.M.Dunn,
and
G.E.Fanucci
(2009).
Subtype polymorphisms among HIV-1 protease variants confer altered flap conformations and flexibility.
|
| |
J Am Chem Soc,
131,
14650-14651.
|
 |
|
|
|
|
 |
P.M.Colman
(2009).
New antivirals and drug resistance.
|
| |
Annu Rev Biochem,
78,
95.
|
 |
|
|
|
|
 |
S.Kazemi,
D.M.Krüger,
F.Sirockin,
and
H.Gohlke
(2009).
Elastic potential grids: accurate and efficient representation of intermolecular interactions for fully flexible docking.
|
| |
ChemMedChem,
4,
1264-1268.
|
 |
|
|
|
|
 |
D.Imamura,
R.Zhou,
M.Feig,
and
L.Kroos
(2008).
Evidence that the Bacillus subtilis SpoIIGA protein is a novel type of signal-transducing aspartic protease.
|
| |
J Biol Chem,
283,
15287-15299.
|
 |
|
|
|
|
 |
F.Liu,
A.Y.Kovalevsky,
Y.Tie,
A.K.Ghosh,
R.W.Harrison,
and
I.T.Weber
(2008).
Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir.
|
| |
J Mol Biol,
381,
102-115.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
G.Verkhivker,
G.Tiana,
C.Camilloni,
D.Provasi,
and
R.A.Broglia
(2008).
Atomistic simulations of the HIV-1 protease folding inhibition.
|
| |
Biophys J,
95,
550-562.
|
 |
|
|
|
|
 |
J.Böttcher,
A.Blum,
S.Dörr,
A.Heine,
W.E.Diederich,
and
G.Klebe
(2008).
Targeting the open-flap conformation of HIV-1 protease with pyrrolidine-based inhibitors.
|
| |
ChemMedChem,
3,
1337-1344.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
M.J.Giffin,
H.Heaslet,
A.Brik,
Y.C.Lin,
G.Cauvi,
C.H.Wong,
D.E.McRee,
J.H.Elder,
C.D.Stout,
and
B.E.Torbett
(2008).
A copper(I)-catalyzed 1,2,3-triazole azide-alkyne click compound is a potent inhibitor of a multidrug-resistant HIV-1 protease variant.
|
| |
J Med Chem,
51,
6263-6270.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
code is
shown on the right.
|
 |

