 |
PDBsum entry 2p1c

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.5.1.10
- (2E,6E)-farnesyl diphosphate synthase.
|
|
 |
 |
 |
 |
 |

Pathway:
|
 |
Terpenoid biosynthesis
|
 |
 |
 |
 |
 |
Reaction:
|
 |
isopentenyl diphosphate + (2E)-geranyl diphosphate = (2E,6E)-farnesyl diphosphate + diphosphate
|
 |
 |
 |
 |
 |
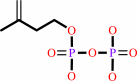 isopentenyl diphosphate
isopentenyl diphosphate
|
+
|
(2E)-geranyl diphosphate
|
=
|
 (2E,6E)-farnesyl diphosphate
(2E,6E)-farnesyl diphosphate
|
+
|
 diphosphate
diphosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proteins
73:431-439
(2008)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structures of a potent phenylalkyl bisphosphonate inhibitor bound to farnesyl and geranylgeranyl diphosphate synthases.
|
|
R.Cao,
C.K.Chen,
R.T.Guo,
A.H.Wang,
E.Oldfield.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
We report the X-ray crystallographic structures of the bisphosphonate
N-[methyl(4-phenylbutyl)]-3-aminopropyl-1-hydroxy-1,1-bisphosphonate (BPH-210),
a potent analog of pamidronate (Aredia), bound to farnesyl diphosphate synthase
(FPPS) from Trypanosoma brucei as well as to geranylgeranyl diphosphate synthase
from Saccharomyces cerevisiae. BPH-210 binds to FPPS, together with 3 Mg(2+),
with its long, hydrophobic phenylbutyl side-chain being located in the same
binding pocket that is occupied by allylic diphosphates and other
bisphosphonates. Binding is overwhelmingly entropy driven, as determined by
isothermal titration calorimetry. The structure is of interest since it explains
the lack of potency of longer chain analogs against FPPS, since these would be
expected to have a steric clash with an aromatic ring at the distal end of the
binding site. Unlike shorter chain FPPS inhibitors, such as pamidronate, BPH-210
is also found to be a potent inhibitor of human geranylgeranyl diphosphate
synthase. In this case, the bisphosphonate binds only to the GGPP product
inhibitory site, with only 1 (chain A) or 0 (chain B) Mg(2+), and DeltaS is much
smaller and DeltaH is approximately 6 k cal more negative than in the case of
FPPS binding. Overall, these results are of general interest since they show
that some bisphosphonates can bind to more than one trans-prenyl synthase enzyme
which, in some cases, can be expected to enhance their overall activity in vitro
and in vivo. Proteins 2008. (c) 2008 Wiley-Liss, Inc.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Figure 1. Structure and interactions in FPPS·BPH-210.
(A) Schematic of T. brucei FPPS·BPH-210 dimer. (B)
Ligplot interaction diagram show FPPS·BPH-210
interactions in chain A. (C) Ligplot interaction diagram of
FPPS·BPH-210 interactions in chain B. (D) 2Fo-Fc electron
density map for BPH-210 bound to FPPS in chain A (green
contoured at 1  ,
red at 3 ,
red at 3  ).
(E) 2Fo-Fc electron density map for BPH-210 in chain B (green
contoured at 1 ).
(E) 2Fo-Fc electron density map for BPH-210 in chain B (green
contoured at 1  ,
red at 3 ,
red at 3  ). ).
|
 |
Figure 3.
Figure 3. Partial sequence alignment of rat, human, chicken, T.
brucei, T. gondi, FPPSs. The  first
DDXXD first
DDXXD  repeat
is shaded in red, the two aromatic amino-acids 4 and 5 residues
upstream of the first DDXXD repeat (PhePhe in rat, human and
chicken, HisTyr in T. brucei) that control to a significant
extent, product specificity, and which are likely to interact
with the phenyl ring in BPH-210, are shown in green. repeat
is shaded in red, the two aromatic amino-acids 4 and 5 residues
upstream of the first DDXXD repeat (PhePhe in rat, human and
chicken, HisTyr in T. brucei) that control to a significant
extent, product specificity, and which are likely to interact
with the phenyl ring in BPH-210, are shown in green.
|
 |
|
|

|
| |
The above figures are
reprinted
from an Open Access publication published by John Wiley & Sons, Inc.:
Proteins
(2008,
73,
431-439)
copyright 2008.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
V.S.Rosso,
S.H.Szajnman,
L.Malayil,
M.Galizzi,
S.N.Moreno,
R.Docampo,
and
J.B.Rodriguez
(2011).
Synthesis and biological evaluation of new 2-alkylaminoethyl-1,1-bisphosphonic acids against Trypanosoma cruzi and Toxoplasma gondii targeting farnesyl diphosphate synthase.
|
| |
Bioorg Med Chem,
19,
2211-2217.
|
 |
|
|
|
|
 |
R.J.Falconer,
A.Penkova,
I.Jelesarov,
and
B.M.Collins
(2010).
Survey of the year 2008: applications of isothermal titration calorimetry.
|
| |
J Mol Recognit,
23,
395-413.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

