 |
PDBsum entry 2ok4

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2ok4
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|

|

|

|

|

|
 110 a.a.
110 a.a.
|
 |

|

|

|

|

|

|

|
 122 a.a.
122 a.a.
|
 |

|

|

|

|

|

|

|

 360 a.a.
360 a.a.
|
 |

|

|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of aromatic amine dehydrogenase ttq- phenylacetaldehyde adduct oxidized with ferricyanide
|
|
Structure:
|
 |
Aromatic amine dehydrogenase, small subunit. Chain: d, h. Fragment: (residues: 48-182). Aromatic amine dehydrogenase, large subunit. Chain: a, b. Fragment: (residues: 73-433). Ec: 1.4.99.4
|
|
Source:
|
 |
Alcaligenes faecalis. Organism_taxid: 511. Organism_taxid: 511
|
|
Resolution:
|
 |
|
1.45Å
|
R-factor:
|
0.156
|
R-free:
|
0.188
|
|
|
Authors:
|
 |
A.Roujeinikova,D.Leys
|
Key ref:

|
 |
A.Roujeinikova
et al.
(2007).
New insights into the reductive half-reaction mechanism of aromatic amine dehydrogenase revealed by reaction with carbinolamine substrates.
J Biol Chem,
282,
23766-23777.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
16-Jan-07
|
Release date:
|
01-May-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P84887
(AAUA_ALCFA) -
Aralkylamine dehydrogenase light chain from Alcaligenes faecalis
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
182 a.a.
110 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains D, H, A, B:
E.C.1.4.9.2
- aralkylamine dehydrogenase (azurin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
an aralkylamine + 2 oxidized [azurin] + H2O = an aromatic aldehyde + 2 reduced [azurin] + NH4+ + 2 H+
|
 |
 |
 |
 |
 |
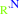 aralkylamine
aralkylamine
|
+
|
2
×
oxidized [azurin]
|
+
|
H2O
|
=
|
aromatic aldehyde
Bound ligand (Het Group name = )
matches with 88.89% similarity
|
+
|
2
×
reduced [azurin]
|
+
|
NH4(+)
|
+
|
2
×
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
282:23766-23777
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
New insights into the reductive half-reaction mechanism of aromatic amine dehydrogenase revealed by reaction with carbinolamine substrates.
|
|
A.Roujeinikova,
P.Hothi,
L.Masgrau,
M.J.Sutcliffe,
N.S.Scrutton,
D.Leys.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Aromatic amine dehydrogenase uses a tryptophan tryptophylquinone (TTQ) cofactor
to oxidatively deaminate primary aromatic amines. In the reductive
half-reaction, a proton is transferred from the substrate C1 to betaAsp-128 O-2,
in a reaction that proceeds by H-tunneling. Using solution studies, kinetic
crystallography, and computational simulation we show that the mechanism of
oxidation of aromatic carbinolamines is similar to amine oxidation, but that
carbinolamine oxidation occurs at a substantially reduced rate. This has enabled
us to determine for the first time the structure of the intermediate prior to
the H-transfer/reduction step. The proton-betaAsp-128 O-2 distance is
approximately 3.7A, in contrast to the distance of approximately 2.7A predicted
for the intermediate formed with the corresponding primary amine substrate. This
difference of approximately 1.0 A is due to an unexpected conformation of the
substrate moiety, which is supported by molecular dynamic simulations and
reflected in the approximately 10(7)-fold slower TTQ reduction rate with
phenylaminoethanol compared with that with primary amines. A water molecule is
observed near TTQ C-6 and is likely derived from the collapse of the preceding
carbinolamine TTQ-adduct. We suggest this water molecule is involved in
consecutive proton transfers following TTQ reduction, and is ultimately
repositioned near the TTQ O-7 concomitant with protein rearrangement. For all
carbinolamines tested, highly stable amide-TTQ adducts are formed following
proton abstraction and TTQ reduction. Slow hydrolysis of the amide occurs after,
rather than prior to, TTQ oxidation and leads ultimately to a carboxylic acid
product.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
FIGURE 3. Schematic overview of the proposed mechanism for
AADH R-carbinolamine oxidation. For ease of comparison with the
previously proposed amine oxidation mechanism, a similar
notation is used according to Fig. 1. For clarity, only part of
the TTQ cofactor is represented, whereas the side chain of the
different R-carbinolamines is indicated by an R. Refer to
supplementary Scheme 1 for a full description of the
substrate-TTQ-enzyme adduct. The active site water molecule (or
ammonia in case of a steady state mechanism, see Ref. 24) is
denoted W1. Whether a conformational equilibrium between IIIb-A
and IIIb-B occurs depends on the nature of the R side chain.
|
 |
Figure 5.
FIGURE 5. Top, comparison of IIIb-A conformation as
observed in the crystals (phenylacetaldehyde derived carbon
atoms in cyan) with a more optimal configuration (IIIb-B) based
on the modeled intermediate IIIa (Fig. 1) during tryptamine
reduction (Ref. 8; phenylacetaldehyde-derived carbon atoms in
yellow). Putative hydrogen bonding interactions made between
IIIb-B conformation and active site residues are shown by dotted
lines. Key active site residues and TTQ cofactor are displayed
with green carbons. B, overlay of crystal structures of IIIb-A
(with green carbons) and Vc (with cyan carbons) for
phenylacetaldehyde and ammonia as substrates. The active site
water molecule situated close to C-6 and O-7 is shown as a red
sphere (labeled W-1[ox]and W-1[red], respectively).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(2007,
282,
23766-23777)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.Pang,
N.S.Scrutton,
S.P.de Visser,
and
M.J.Sutcliffe
(2010).
New insights into the multi-step reaction pathway of the reductive half-reaction catalysed by aromatic amine dehydrogenase: a QM/MM study.
|
| |
Chem Commun (Camb),
46,
3104-3106.
|
 |
|
|
|
|
 |
P.Hothi,
S.Hay,
A.Roujeinikova,
M.J.Sutcliffe,
M.Lee,
D.Leys,
P.M.Cullis,
and
N.S.Scrutton
(2008).
Driving force analysis of proton tunnelling across a reactivity series for an enzyme-substrate complex.
|
| |
Chembiochem,
9,
2839-2845.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |
| |

