 |
PDBsum entry 2i1v

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Transferase, hydrolase
|
PDB id
|
|

|

|
2i1v
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Transferase, hydrolase
|
 |
|
Title:
|
 |
Crystal structure of pfkfb3 in complex with adp and fructose-2,6- bisphosphate
|
|
Structure:
|
 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3. Chain: b. Synonym: pfkfb3, phosphoryl transferase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562
|
|
Resolution:
|
 |
|
2.50Å
|
R-factor:
|
0.214
|
R-free:
|
0.262
|
|
|
Authors:
|
 |
S.G.Kim,M.R.El-Maghrabi,Y.H.Lee
|
Key ref:

|
 |
S.G.Kim
et al.
(2007).
A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase.
J Mol Biol,
370,
14-26.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
15-Aug-06
|
Release date:
|
03-Jul-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q16875
(F263_HUMAN) -
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
520 a.a.
449 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class 2:
|
 |
E.C.2.7.1.105
- 6-phosphofructo-2-kinase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 6-phosphate + ATP = beta-D-fructose 2,6-bisphosphate + ADP + H+
|
 |
 |
 |
 |
 |
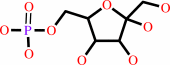 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
+
|
 ATP
ATP
|
=
|
beta-D-fructose 2,6-bisphosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
ADP
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Enzyme class 3:
|
 |
E.C.3.1.3.46
- fructose-2,6-bisphosphate 2-phosphatase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
beta-D-fructose 2,6-bisphosphate + H2O = beta-D-fructose 6-phosphate + phosphate
|
 |
 |
 |
 |
 |
beta-D-fructose 2,6-bisphosphate
Bound ligand (Het Group name = )
corresponds exactly
|
+
|
H2O
|
=
|
 beta-D-fructose 6-phosphate
beta-D-fructose 6-phosphate
|
+
|
phosphate
Bound ligand (Het Group name = )
matches with 80.00% similarity
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Note, where more than one E.C. class is given (as above), each may
correspond to a different protein domain or, in the case of polyprotein
precursors, to a different mature protein.
|
|
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
370:14-26
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
A Direct Substrate-Substrate Interaction Found in the Kinase Domain of the Bifunctional Enzyme, 6-Phosphofructo-2-kinase/Fructose-2,6-bisphosphatase.
|
|
S.G.Kim,
M.Cavalier,
M.R.El-Maghrabi,
Y.H.Lee.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
To understand the molecular basis of a phosphoryl transfer reaction catalyzed by
the 6-phosphofructo-2-kinase domain of the hypoxia-inducible bifunctional enzyme
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase (PFKFB3), the crystal
structures of PFKFB3AMPPCPfructose-6-phosphate and PFKFB3ADPphosphoenolpyruvate
complexes were determined to 2.7 A and 2.25 A resolution, respectively. Kinetic
studies on the wild-type and site-directed mutant proteins were carried out to
confirm the structural observations. The experimentally varied liganding states
in the active pocket cause no significant conformational changes. In the
pseudo-substrate complex, a strong direct interaction between AMPPCP and
fructose-6-phosphate (Fru-6-P) is found. By virtue of this direct
substrate-substrate interaction, Fru-6-P is aligned with AMPPCP in an
orientation and proximity most suitable for a direct transfer of the
gamma-phosphate moiety to 2-OH of Fru-6-P. The three key atoms involved in the
phosphoryl transfer, the beta,gamma-phosphate bridge oxygen atom, the
gamma-phosphorus atom, and the 2-OH group are positioned in a single line,
suggesting a direct phosphoryl transfer without formation of a phosphoenzyme
intermediate. In addition, the distance between 2-OH and gamma-phosphorus allows
the gamma-phosphate oxygen atoms to serve as a general base catalyst to induce
an "associative" phosphoryl transfer mechanism. The site-directed
mutant study and inhibition kinetics suggest that this reaction will be
catalyzed most efficiently by the protein when the substrates bind to the active
pocket in an ordered manner in which ATP binds first.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. A cartoon of the suggested catalytic pathway. In
the clockwise direction, 2-OH of Fru-6-P is deprotonated by the
γ-phosphate moiety of ATP and a nucleophilic attack occurs. An
intermediate pentavalent phosphorane is formed, and the negative
charges generated on the planary oxygen atoms are stabilized by
Lys47, Lys168, and Mg^2+, and maybe a H^+ extracted from 2-OH.
Redistribution of the phosphorane electrons breaks a diester
bond between the bridge oxygen and γ-phosphorus atoms. The
leaving group is stabilized by a H^+. Eventually, the leaving
group (ADP) is stabilized through a salt-bridge to Lys168 to
finish the reaction.
|
 |
Figure 4.
Figure 4. PEP binding to PFKFB3. (a) The |F[o]|–|F[c]| omit
electron density map. The map is calculated in the absence of
ligands and contoured at 2.5σ. (b) A stereo view of the
interactions of PEP with the 2-Kase active site pocket is shown.
The dotted lines represent hydrogen bonds or salt-bridges. To
show its position in the 2-Kase active pocket, the structure of
PFKFB3 radical dot AMPPCP radical dot Fru-6-P complex is
superposed and Fru-6-P (light gray) is shown.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2007,
370,
14-26)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

