
|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
Chains A, B, C, D:
E.C.3.5.1.26
- N(4)-(beta-N-acetylglucosaminyl)-L-asparaginase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
N4-(beta-N-acetyl-D-glucosaminyl)-L-asparagine + H2O = N-acetyl-beta-D- glucosaminylamine + L-aspartate + H+
|
 |
 |
 |
 |
 |
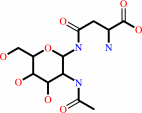 N(4)-(beta-N-acetyl-D-glucosaminyl)-L-asparagine
N(4)-(beta-N-acetyl-D-glucosaminyl)-L-asparagine
|
+
|
H2O
|
=
|
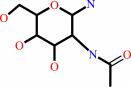 N-acetyl-beta-D- glucosaminylamine
N-acetyl-beta-D- glucosaminylamine
|
+
|
 L-aspartate
L-aspartate
|
+
|
H(+)
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Biol Chem
273:20205-20212
(1998)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structures of Flavobacterium glycosylasparaginase. An N-terminal nucleophile hydrolase activated by intramolecular proteolysis.
|
|
H.C.Guo,
Q.Xu,
D.Buckley,
C.Guan.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Glycosylasparaginase (GA) is a member of a novel family of N-terminal
nucleophile hydrolases that catalytically use an N-terminal residue as both a
polarizing base and a nucleophile. These enzymes are activated from a single
chain precursor by intramolecular autoproteolysis to yield the N-terminal
nucleophile. A deficiency of GA results in the human genetic disorder known as
aspartylglycosaminuria. In this study, we report the crystal structure of
recombinant GA from Flavobacterium meningosepticum. Similar to the human
structure, the bacterial GA forms an alphabetabetaalpha sandwich. However, some
significant differences are observed between the Flavobacterium and human
structures. The active site of Flavobacterium glycosylasparaginase is in an open
conformation when compared with the human structure. We also describe the
structure of a mutant wherein the N-terminal nucleophile Thr152 is substituted
by a cysteine. In the bacterial GA crystals, we observe a heterotetrameric
structure similar to that found in the human structure, as well as that observed
in solution for eukaryotic glycosylasparaginases. The results confirm the
suitability of the bacterial enzyme as a model to study the consequences of
mutations in aspartylglycosaminuria patients. They also suggest that further
studies are necessary to understand the detail mechanism of this enzyme. The
presence of the heterotetrameric structure in the crystals is significant
because dimerization of precursors has been suggested in the human enzyme to be
a prerequisite to trigger autoproteolysis.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. The structure of glycosylasparaginase from F.
meningosepticum. a, stereo ribbon representation of the
Flavobacterium GA structure. One heterodimer is shown with a-
(red) and b-subunits (green). The active site is at top center
of the structure toward the viewer and around the N-terminal end
of the b-subunit (green) (labeled Nb in light blue). b, stereo
diagram of C  traces of
Flavobacterium GA. c, stereo diagram of C traces of
Flavobacterium GA. c, stereo diagram of C  traces of
Flavobacterium (dark blue) and human (gray) GA. Superimposition
is based on all common main chain atoms (excluding
insertions/deletions). The residues labeled are in
Flavobacterium sequence number. traces of
Flavobacterium (dark blue) and human (gray) GA. Superimposition
is based on all common main chain atoms (excluding
insertions/deletions). The residues labeled are in
Flavobacterium sequence number.
|
 |
Figure 4.
Fig. 4. Stereo view of the active site of
glycosylasparaginase. a, stereo view of superimposition of the
active sites between Flavobacterium (shown according to atom
type: yellow for carbons, blue for nitrogens, and red for
oxygens) and human (shown in gray) GA. Also shown is aspartate
in the human enzyme/product structure (13). Dashed lines
correspond to the hydrogen bonds described in the human
structure. b, the same stereo view of active site in the T152C
mutant. The color scheme is the same as the wild type in (a),
except that sulfur of the thiol group is shown in green.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from the ASBMB:
J Biol Chem
(1998,
273,
20205-20212)
copyright 1998.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
J.R.Cantor,
E.M.Stone,
L.Chantranupong,
and
G.Georgiou
(2009).
The human asparaginase-like protein 1 hASRGL1 is an Ntn hydrolase with beta-aspartyl peptidase activity.
|
| |
Biochemistry,
48,
11026-11031.
|
 |
|
|
|
|
 |
K.Michalska,
D.Borek,
A.Hernández-Santoyo,
and
M.Jaskolski
(2008).
Crystal packing of plant-type L-asparaginase from Escherichia coli.
|
| |
Acta Crystallogr D Biol Crystallogr,
64,
309-320.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Sun,
and
H.C.Guo
(2008).
Structural constraints on autoprocessing of the human nucleoporin Nup98.
|
| |
Protein Sci,
17,
494-505.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.A.Cañas,
F.de la Torre,
F.M.Cánovas,
and
F.R.Cantón
(2007).
Coordination of PsAS1 and PsASPG expression controls timing of re-allocated N utilization in hypocotyls of pine seedlings.
|
| |
Planta,
225,
1205-1219.
|
 |
|
|
|
|
 |
Y.Wang,
and
H.C.Guo
(2007).
Crystallographic snapshot of a productive glycosylasparaginase-substrate complex.
|
| |
J Mol Biol,
366,
82-92.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
L.L.Lin,
P.R.Chou,
Y.W.Hua,
and
W.H.Hsu
(2006).
Overexpression, one-step purification, and biochemical characterization of a recombinant gamma-glutamyltranspeptidase from Bacillus licheniformis.
|
| |
Appl Microbiol Biotechnol,
73,
103-112.
|
 |
|
|
|
|
 |
J.A.Khan,
B.M.Dunn,
and
L.Tong
(2005).
Crystal structure of human Taspase1, a crucial protease regulating the function of MLL.
|
| |
Structure,
13,
1443-1452.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.Prahl,
M.Pazgier,
M.Hejazi,
W.Lockau,
and
J.Lubkowski
(2004).
Structure of the isoaspartyl peptidase with L-asparaginase activity from Escherichia coli.
|
| |
Acta Crystallogr D Biol Crystallogr,
60,
1173-1176.
|
 |
|
PDB code:
|
 |
|
|
|
|
|
 |
D.Borek,
K.Michalska,
K.Brzezinski,
A.Kisiel,
J.Podkowinski,
D.T.Bonthron,
D.Krowarsch,
J.Otlewski,
and
M.Jaskolski
(2004).
Expression, purification and catalytic activity of Lupinus luteus asparagine beta-amidohydrolase and its Escherichia coli homolog.
|
| |
Eur J Biochem,
271,
3215-3226.
|
 |
|
|
|
|
 |
F.Schmitzberger,
M.L.Kilkenny,
C.M.Lobley,
M.E.Webb,
M.Vinkovic,
D.Matak-Vinkovic,
M.Witty,
D.Y.Chirgadze,
A.G.Smith,
C.Abell,
and
T.L.Blundell
(2003).
Structural constraints on protein self-processing in L-aspartate-alpha-decarboxylase.
|
| |
EMBO J,
22,
6193-6204.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
X.Qian,
C.Guan,
and
H.C.Guo
(2003).
A dual role for an aspartic acid in glycosylasparaginase autoproteolysis.
|
| |
Structure,
11,
997.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
R.A.Larsen,
T.M.Knox,
and
C.G.Miller
(2001).
Aspartic peptide hydrolases in Salmonella enterica serovar typhimurium.
|
| |
J Bacteriol,
183,
3089-3097.
|
 |
|
|
|
|
 |
C.Oinonen,
and
J.Rouvinen
(2000).
Structural comparison of Ntn-hydrolases.
|
| |
Protein Sci,
9,
2329-2337.
|
 |
|
|
|
|
 |
D.Borek,
and
M.Jaskólski
(2000).
Crystallization and preliminary crystallographic studies of a new L-asparaginase encoded by the Escherichia coli genome.
|
| |
Acta Crystallogr D Biol Crystallogr,
56,
1505-1507.
|
 |
|
|
|
|
 |
H.Paulus
(2000).
Protein splicing and related forms of protein autoprocessing.
|
| |
Annu Rev Biochem,
69,
447-496.
|
 |
|
|
|
|
 |
M.Inoue,
J.Hiratake,
H.Suzuki,
H.Kumagai,
and
K.Sakata
(2000).
Identification of catalytic nucleophile of Escherichia coli gamma-glutamyltranspeptidase by gamma-monofluorophosphono derivative of glutamic acid: N-terminal thr-391 in small subunit is the nucleophile.
|
| |
Biochemistry,
39,
7764-7771.
|
 |
|
|
|
|
 |
N.N.Aronson
(1999).
Aspartylglycosaminuria: biochemistry and molecular biology.
|
| |
Biochim Biophys Acta,
1455,
139-154.
|
 |
|
|
|
|
 |
Q.Xu,
D.Buckley,
C.Guan,
and
H.C.Guo
(1999).
Structural insights into the mechanism of intramolecular proteolysis.
|
| |
Cell,
98,
651-661.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
S.Li,
J.L.Smith,
and
H.Zalkin
(1999).
Mutational analysis of Bacillus subtilis glutamine phosphoribosylpyrophosphate amidotransferase propeptide processing.
|
| |
J Bacteriol,
181,
1403-1408.
|
 |
|
|
|
|
 |
T.Cui,
P.H.Liao,
C.Guan,
and
H.C.Guo
(1999).
Purification and crystallization of precursors and autoprocessed enzymes of Flavobacterium glycosylasparaginase: an N-terminal nucleophile hydrolase.
|
| |
Acta Crystallogr D Biol Crystallogr,
55,
1961-1964.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

