 |
PDBsum entry 2f8m

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.5.3.1.6
- ribose-5-phosphate isomerase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
aldehydo-D-ribose 5-phosphate = D-ribulose 5-phosphate
|
 |
 |
 |
 |
 |
aldehydo-D-ribose 5-phosphate
|
=
|
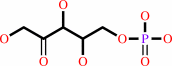 D-ribulose 5-phosphate
D-ribulose 5-phosphate
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Acta Crystallograph Sect F Struct Biol Cryst Commun
62:427-431
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structure of ribose 5-phosphate isomerase from Plasmodium falciparum.
|
|
M.A.Holmes,
F.S.Buckner,
W.C.Van Voorhis,
C.L.Verlinde,
C.Mehlin,
E.Boni,
G.DeTitta,
J.Luft,
A.Lauricella,
L.Anderson,
O.Kalyuzhniy,
F.Zucker,
L.W.Schoenfeld,
T.N.Earnest,
W.G.Hol,
E.A.Merritt.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The structure of ribose 5-phosphate isomerase from Plasmodium falciparum,
PFE0730c, has been determined by molecular replacement at 2.09 angstroms
resolution. The enzyme, which catalyzes the isomerization reaction that
interconverts ribose 5-phosphate and ribulose 5-phosphate, is a member of the
pentose phosphate pathway. The P. falciparum enzyme belongs to the ribose
5-phosphate isomerase A family, Pfam family PF06562 (DUF1124), and is
structurally similar to other members of the family.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |
Figure 1.
Figure 1 Main figure: ribbon representation of the
Pfal008434AAA dimer. The A chain is colored smoothly from blue
at the N-terminus to red at the C-terminus. The B chain is
colored by domain, with the larger domain in pink and the
smaller domain in purple. The tetramerization loop of chain A is
indicated by an arrow. Inset: enlarged view of the active site
of the Pfal008434AAA A chain. The bound phosphate ion is shown
in stick form and the three water molecules mentioned in the
text are indicated as spheres. The substrate ribose 5-phosphate
(light blue) from the T. thermophilus structure 1uj5 is shown
superposed onto the Pfal008434AAA active site based on
superposition of 27 C^  atoms
in two stretches of chain that contain ten of the 11 residues
directly in contact with the ribose 5-phosphate. The figure was
generated using PyMOL (DeLano, 2002[DeLano, W. (2002). The PyMOL
Molecular Graphics System. http://www.pymol.org .]). atoms
in two stretches of chain that contain ten of the 11 residues
directly in contact with the ribose 5-phosphate. The figure was
generated using PyMOL (DeLano, 2002[DeLano, W. (2002). The PyMOL
Molecular Graphics System. http://www.pymol.org .]).
|
 |
|
|

|
| |
The above figure is
reprinted
from an Open Access publication published by the IUCr:
Acta Crystallograph Sect F Struct Biol Cryst Commun
(2006,
62,
427-431)
copyright 2006.
|
|
| |
Figure was
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

