 |
PDBsum entry 2dgd

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Lyase
|
 |
|
Title:
|
 |
Crystal structure of st0656, a function unknown protein from sulfolobus tokodaii
|
|
Structure:
|
 |
223aa long hypothetical arylmalonate decarboxylase. Chain: a, b, c, d. Engineered: yes
|
|
Source:
|
 |
Sulfolobus tokodaii. Organism_taxid: 111955. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.90Å
|
R-factor:
|
0.188
|
R-free:
|
0.222
|
|
|
Authors:
|
 |
Y.Tanaka,T.Sasaki,E.Tanabe,M.Yao,I.Tanaka,I.Kumagai,K.Tsumoto
|
|
Key ref:
|
 |
Y.Tanaka
et al.
Crystal structure of st0656, A function unknown protein from sulfolobus tokodaii.
To be published,
.
|
 |
|
Date:
|
 |
|
10-Mar-06
|
Release date:
|
13-Mar-07
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
Q974K3
(Q974K3_SULTO) -
Arylmalonate decarboxylase from Sulfurisphaera tokodaii (strain DSM 16993 / JCM 10545 / NBRC 100140 / 7)
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
223 a.a.
222 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.4.1.1.76
- arylmalonate decarboxylase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
2-aryl-2-methylmalonate + H+ = 2-arylpropionate + CO2
|
 |
 |
 |
 |
 |
2-aryl-2-methylmalonate
Bound ligand (Het Group name = )
matches with 50.00% similarity
|
+
|
H(+)
|
=
|
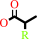 2-arylpropionate
2-arylpropionate
|
+
|
 CO2
CO2
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Divalent cation; Thiamine diphosphate
|
 |
 |
 |
 |
 |
Divalent cation
|
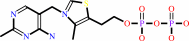 Thiamine diphosphate
Thiamine diphosphate
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |

