 |
PDBsum entry 2d51

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
* Residue conservation analysis
|
|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.2.3.1.216
- 5,7-dihydroxy-2-methylchromone synthase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
5 malonyl-CoA + 4 H+ = 5,7-dihydroxy-2-methyl-4H-chromen-4-one + 5 CO2 + 5 CoA + H2O
|
 |
 |
 |
 |
 |
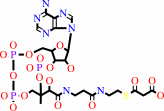 5
×
malonyl-CoA
5
×
malonyl-CoA
|
+
|
4
×
H(+)
|
=
|
5,7-dihydroxy-2-methyl-4H-chromen-4-one
|
+
|
 5
×
CO2
5
×
CO2
|
+
|
 5
×
CoA
5
×
CoA
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Chem Biol
14:359-369
(2007)
|
|
PubMed id:
|
|
|
|

|
| |
|
Structural insight into chain-length control and product specificity of pentaketide chromone synthase from Aloe arborescens.
|
|
H.Morita,
S.Kondo,
S.Oguro,
H.Noguchi,
S.Sugio,
I.Abe,
T.Kohno.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
The crystal structures of a wild-type and a mutant PCS, a novel plant type III
polyketide synthase from a medicinal plant, Aloe arborescens, were solved at 1.6
A resolution. The crystal structures revealed that the pentaketide-producing
wild-type and the octaketide-producing M207G mutant shared almost the same
overall folding, and that the large-to-small substitution dramatically increases
the volume of the polyketide-elongation tunnel by opening a gate to two hidden
pockets behind the active site of the enzyme. The chemically inert active site
residue 207 thus controls the number of condensations of malonyl-CoA, solely
depending on the steric bulk of the side chain. These findings not only provided
insight into the polyketide formation reaction, but they also suggested
strategies for the engineered biosynthesis of polyketides.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 3.
Figure 3. Overall Structure of PCS Complexed with CoA-SH
(A) Ribbon representation of the PCS homodimer. The monomers are
colored green and silver, and the CoA-SH molecules are shown as
blue stick models. The catalytic Cys174 and Met147, which form a
partial wall of the active-site cavity of another monomer, are
highlighted as yellow CPK and stick models, respectively.
(B) Comparison of PCS (green), M. sativa CHS (blue), and G.
hybrida 2PS (purple). The catalytic Cys174 and the bound CoA-SH
in PCS are also shown as yellow and red CPK molecules,
respectively.
(C) CoA-SH binding to the PCS structure. The
CoA-SH (green) and the SIGMA-weighted |2F[o] − F[c]| electron
density (0.8σ, red cage) for CoA-SH are shown. The water
molecules (light-blue spheres) and hydrogen bonds (dotted lines)
are also indicated.
|
 |
Figure 6.
Figure 6. Schematic Representation of the Active-Site
Architecture of Wild-Type PCS, the M207G Mutant, and M. sativa
CHS
(A–C) The M207G substitution opens a gate to the
buried pocket A that extends into the “floor” of the
active-site cavity, resulting in a 4:1 mixture of SEK4b:SEK4
instead of 5,7-dihydroxy- 2-methylchormone. PCS locks the methyl
end of its linear pentaketide intermediate between Met207 and
Val351, as in the case in which M. sativa CHS locks the aromatic
ring derived from 4-coumaroyl-CoA with the coumaroyl-binding
pocket.
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Cell Press:
Chem Biol
(2007,
14,
359-369)
copyright 2007.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
D.Cook,
A.M.Rimando,
T.E.Clemente,
J.Schröder,
F.E.Dayan,
N.P.Nanayakkara,
Z.Pan,
B.P.Noonan,
M.Fishbein,
I.Abe,
S.O.Duke,
and
S.R.Baerson
(2010).
Alkylresorcinol synthases expressed in Sorghum bicolor root hairs play an essential role in the biosynthesis of the allelopathic benzoquinone sorgoleone.
|
| |
Plant Cell,
22,
867-887.
|
 |
|
|
|
|
 |
H.Morita,
Y.Shimokawa,
M.Tanio,
R.Kato,
H.Noguchi,
S.Sugio,
T.Kohno,
and
I.Abe
(2010).
A structure-based mechanism for benzalacetone synthase from Rheum palmatum.
|
| |
Proc Natl Acad Sci U S A,
107,
669-673.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
I.Abe,
and
H.Morita
(2010).
Structure and function of the chalcone synthase superfamily of plant type III polyketide synthases.
|
| |
Nat Prod Rep,
27,
809-838.
|
 |
|
|
|
|
 |
P.K.Koduri,
G.S.Gordon,
E.I.Barker,
C.C.Colpitts,
N.W.Ashton,
and
D.Y.Suh
(2010).
Genome-wide analysis of the chalcone synthase superfamily genes of Physcomitrella patens.
|
| |
Plant Mol Biol,
72,
247-263.
|
 |
|
|
|
|
 |
I.Fujii
(2009).
Heterologous expression systems for polyketide synthases.
|
| |
Nat Prod Rep,
26,
155-169.
|
 |
|
|
|
|
 |
T.Klundt,
M.Bocola,
M.Lütge,
T.Beuerle,
B.Liu,
and
L.Beerhues
(2009).
A single amino acid substitution converts benzophenone synthase into phenylpyrone synthase.
|
| |
J Biol Chem,
284,
30957-30964.
|
 |
|
|
|
|
 |
Y.Mizuuchi,
S.P.Shi,
K.Wanibuchi,
A.Kojima,
H.Morita,
H.Noguchi,
and
I.Abe
(2009).
Novel type III polyketide synthases from Aloe arborescens.
|
| |
FEBS J,
276,
2391-2401.
|
 |
|
|
|
|
 |
C.Taguchi,
F.Taura,
T.Tamada,
Y.Shoyama,
Y.Shoyama,
H.Tanaka,
R.Kuroki,
and
S.Morimoto
(2008).
Crystallization and preliminary X-ray diffraction studies of polyketide synthase-1 (PKS-1) from Cannabis sativa.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
217-220.
|
 |
|
|
|
|
 |
H.Morita,
M.Tanio,
S.Kondo,
R.Kato,
K.Wanibuchi,
H.Noguchi,
S.Sugio,
I.Abe,
and
T.Kohno
(2008).
Crystallization and preliminary crystallographic analysis of a plant type III polyketide synthase that produces benzalacetone.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
64,
304-306.
|
 |
|
|
|
|
 |
I.Abe
(2008).
Engineering of plant polyketide biosynthesis.
|
| |
Chem Pharm Bull (Tokyo),
56,
1505-1514.
|
 |
|
|
|
|
 |
O.Yu,
and
J.M.Jez
(2008).
Nature's assembly line: biosynthesis of simple phenylpropanoids and polyketides.
|
| |
Plant J,
54,
750-762.
|
 |
|
|
|
|
 |
S.B.Rubin-Pitel,
H.Zhang,
T.Vu,
J.S.Brunzelle,
H.Zhao,
and
S.K.Nair
(2008).
Distinct structural elements dictate the specificity of the type III pentaketide synthase from Neurospora crassa.
|
| |
Chem Biol,
15,
1079-1090.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
Y.Mizuuchi,
Y.Shimokawa,
K.Wanibuchi,
H.Noguchi,
and
I.Abe
(2008).
Structure function analysis of novel type III polyketide synthases from Arabidopsis thaliana.
|
| |
Biol Pharm Bull,
31,
2205-2210.
|
 |
|
|
|
|
 |
H.Morita,
S.Kondo,
R.Kato,
K.Wanibuchi,
H.Noguchi,
S.Sugio,
I.Abe,
and
T.Kohno
(2007).
Crystallization and preliminary crystallographic analysis of an acridone-producing novel multifunctional type III polyketide synthase from Huperzia serrata.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
576-578.
|
 |
|
|
|
|
 |
H.Morita,
S.Kondo,
R.Kato,
K.Wanibuchi,
H.Noguchi,
S.Sugio,
I.Abe,
and
T.Kohno
(2007).
Crystallization and preliminary crystallographic analysis of an octaketide-producing plant type III polyketide synthase.
|
| |
Acta Crystallogr Sect F Struct Biol Cryst Commun,
63,
947-949.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

