 |
PDBsum entry 2d0t

|
|
|

|
 |

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
 |

|

|

|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|

|

|

|

|

|

|
|
Oxidoreductase
|
PDB id
|
|

|

|
2d0t
|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Oxidoreductase
|
 |
|
Title:
|
 |
Crystal structure of 4-phenylimidazole bound form of human indoleamine 2,3-dioxygenase
|
|
Structure:
|
 |
Indoleamine 2,3-dioxygenase. Chain: a, b. Synonym: ido, indoleamine-pyrrole 2,3-dioxygenase. Engineered: yes
|
|
Source:
|
 |
Homo sapiens. Human. Organism_taxid: 9606. Expressed in: escherichia coli. Expression_system_taxid: 562.
|
|
Resolution:
|
 |
|
2.30Å
|
R-factor:
|
0.191
|
R-free:
|
0.221
|
|
|
Authors:
|
 |
H.Sugimoto,S.Oda,T.Otsuki,T.Hino,T.Yoshida,Y.Shiro,Riken Structural Genomics/proteomics Initiative (Rsgi)
|
Key ref:

|
 |
H.Sugimoto
et al.
(2006).
Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
Proc Natl Acad Sci U S A,
103,
2611-2616.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
08-Aug-05
|
Release date:
|
31-Jan-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P14902
(I23O1_HUMAN) -
Indoleamine 2,3-dioxygenase 1 from Homo sapiens
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
403 a.a.
373 a.a.
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.13.11.52
- indoleamine 2,3-dioxygenase.
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
|
1.
|
D-tryptophan + O2 = N-formyl-D-kynurenine
|
|
2.
|
L-tryptophan + O2 = N-formyl-L-kynurenine
|
|
 |
 |
 |
 |
 |
D-tryptophan
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
 O2
O2
|
=
|
 N-formyl-D-kynurenine
N-formyl-D-kynurenine
|
|
 |
 |
 |
 |
 |
L-tryptophan
Bound ligand (Het Group name = )
matches with 62.50% similarity
|
+
|
 O2
O2
|
=
|
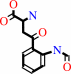 N-formyl-L-kynurenine
N-formyl-L-kynurenine
|
|
 |
 |
 |
 |
 |
 |
 |
 |
Cofactor:
|
 |
Heme
|
 |
 |
 |
 |
 |
Heme
Bound ligand (Het Group name =
HEM)
matches with 95.45% similarity
|
|
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
Proc Natl Acad Sci U S A
103:2611-2616
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Crystal structure of human indoleamine 2,3-dioxygenase: catalytic mechanism of O2 incorporation by a heme-containing dioxygenase.
|
|
H.Sugimoto,
S.Oda,
T.Otsuki,
T.Hino,
T.Yoshida,
Y.Shiro.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
Human indoleamine 2,3-dioxygenase (IDO) catalyzes the cleavage of the pyrrol
ring of L-Trp and incorporates both atoms of a molecule of oxygen (O2). Here we
report on the x-ray crystal structure of human IDO, complexed with the ligand
inhibitor 4-phenylimidazole and cyanide. The overall structure of IDO shows two
alpha-helical domains with the heme between them. A264 of the flexible loop in
the heme distal side is in close proximity to the iron. A mutant analysis shows
that none of the polar amino acid residues in the distal heme pocket are
essential for activity, suggesting that, unlike the heme-containing
monooxygenases (i.e., peroxidase and cytochrome P450), no protein group of IDO
is essential in dioxygen activation or proton abstraction. These characteristics
of the IDO structure provide support for a reaction mechanism involving the
abstraction of a proton from the substrate by iron-bound dioxygen. Inactive
mutants (F226A, F227A, and R231A) retain substrate-binding affinity, and an
electron density map reveals that 2-(N-cyclohexylamino)ethane sulfonic acid is
bound to these residues, mimicking the substrate. These findings suggest that
strict shape complementarities between the indole ring of the substrate and the
protein side chains are required, not for binding, but, rather, to permit the
interaction between the substrate and iron-bound dioxygen in the first step of
the reaction. This study provides the structural basis for a heme-containing
dioxygenase mechanism, a missing piece in our understanding of heme chemistry.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 1.
Fig. 1. Structure of IDO–PI complex. (A) Ribbon
representation of the overall structure of human IDO. The small
and large domains are represented by blue and green ribbons,
respectively. The helices A–S are named in the order of
appearance in the primary sequence. The connecting helices (K-L
and N) are colored in cyan. The long loop connecting the two
domains is colored in red. The heme (yellow), proximal ligand
H346 (white), and heme inhibitor 4-phynylimidazole (white) are
shown in a ball-and-stick model. The helices of the large domain
create the cavity for the heme. The connecting loop (red) and
small domain above the sixth-coordination site (heme distal
side) cover the top of cavity on the heme. (B) The four proximal
helices I, G, Q, and S run in parallel. The helices N (blue) and
K-L (cyan) connect the two domains. The connecting loop (red)
and small domain above the sixth-coordination site of the heme
cover the top of the cavity on the heme.
|
 |
Figure 3.
Fig. 3. Active site of IDO–PI complex. (A) Stereoview of
the residues around the heme of IDO viewed from the side of heme
plane. The proximal ligand H346 is H-bonded to wa1. The
6-propionate of the heme contacts with wa2 and R343 N  . The
wa2 is H-bonded to wa1, L388 O, and 6-propionate. Mutations of
F226, F227, and R231 do not lose the substrate affinity but
produce the inactive enzyme. Two CHES molecules are bound in the
distal pocket. The cyclohexan ring of CHES-1 (green) contacts
with F226 and R231. The 7-propionate of the heme interacts with
the amino group of CHES-1 and side chain of Ser-263. The
mutational analyses for these distal residues are shown in Table
1. (B) Top view of A by a rotation of 90°. The proximal
residues are omitted. . The
wa2 is H-bonded to wa1, L388 O, and 6-propionate. Mutations of
F226, F227, and R231 do not lose the substrate affinity but
produce the inactive enzyme. Two CHES molecules are bound in the
distal pocket. The cyclohexan ring of CHES-1 (green) contacts
with F226 and R231. The 7-propionate of the heme interacts with
the amino group of CHES-1 and side chain of Ser-263. The
mutational analyses for these distal residues are shown in Table
1. (B) Top view of A by a rotation of 90°. The proximal
residues are omitted.
|
 |
|
|
| |
Figures were
selected
by the author.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
K.Nienhaus,
E.Nickel,
C.Lu,
S.R.Yeh,
and
G.U.Nienhaus
(2011).
Ligand migration in human indoleamine-2,3 dioxygenase.
|
| |
IUBMB Life,
63,
153-159.
|
 |
|
|
|
|
 |
C.J.Austin,
B.M.Mailu,
G.J.Maghzal,
A.Sanchez-Perez,
S.Rahlfs,
K.Zocher,
H.J.Yuasa,
J.W.Arthur,
K.Becker,
R.Stocker,
N.H.Hunt,
and
H.J.Ball
(2010).
Biochemical characteristics and inhibitor selectivity of mouse indoleamine 2,3-dioxygenase-2.
|
| |
Amino Acids,
39,
565-578.
|
 |
|
|
|
|
 |
F.Hofmann
(2010).
Ido brings down the pressure in systemic inflammation.
|
| |
Nat Med,
16,
265-267.
|
 |
|
|
|
|
 |
L.Capece,
A.Lewis-Ballester,
D.Batabyal,
N.Di Russo,
S.R.Yeh,
D.A.Estrin,
and
M.A.Marti
(2010).
The first step of the dioxygenation reaction carried out by tryptophan dioxygenase and indoleamine 2,3-dioxygenase as revealed by quantum mechanical/molecular mechanical studies.
|
| |
J Biol Inorg Chem,
15,
811-823.
|
 |
|
|
|
|
 |
L.Capece,
M.Arrar,
A.E.Roitberg,
S.R.Yeh,
M.A.Marti,
and
D.A.Estrin
(2010).
Substrate stereo-specificity in tryptophan dioxygenase and indoleamine 2,3-dioxygenase.
|
| |
Proteins,
78,
2961-2972.
|
 |
|
|
|
|
 |
L.Huang,
B.Baban,
B.A.Johnson,
and
A.L.Mellor
(2010).
Dendritic cells, indoleamine 2,3 dioxygenase and acquired immune privilege.
|
| |
Int Rev Immunol,
29,
133-155.
|
 |
|
|
|
|
 |
L.J.Smith,
A.Kahraman,
and
J.M.Thornton
(2010).
Heme proteins--diversity in structural characteristics, function, and folding.
|
| |
Proteins,
78,
2349-2368.
|
 |
|
|
|
|
 |
R.M.Davydov,
N.Chauhan,
S.J.Thackray,
J.L.Anderson,
N.D.Papadopoulou,
C.G.Mowat,
S.K.Chapman,
E.L.Raven,
and
B.M.Hoffman
(2010).
Probing the ternary complexes of indoleamine and tryptophan 2,3-dioxygenases by cryoreduction EPR and ENDOR spectroscopy.
|
| |
J Am Chem Soc,
132,
5494-5500.
|
 |
|
|
|
|
 |
U.Grohmann,
and
V.Bronte
(2010).
Control of immune response by amino acid metabolism.
|
| |
Immunol Rev,
236,
243-264.
|
 |
|
|
|
|
 |
A.Lewis-Ballester,
D.Batabyal,
T.Egawa,
C.Lu,
Y.Lin,
M.A.Marti,
L.Capece,
D.A.Estrin,
and
S.R.Yeh
(2009).
Evidence for a ferryl intermediate in a heme-based dioxygenase.
|
| |
Proc Natl Acad Sci U S A,
106,
17371-17376.
|
 |
|
|
|
|
 |
A.Macchiarulo,
E.Camaioni,
R.Nuti,
and
R.Pellicciari
(2009).
Highlights at the gate of tryptophan catabolism: a review on the mechanisms of activation and regulation of indoleamine 2,3-dioxygenase (IDO), a novel target in cancer disease.
|
| |
Amino Acids,
37,
219-229.
|
 |
|
|
|
|
 |
B.A.Johnson,
B.Baban,
and
A.L.Mellor
(2009).
Targeting the immunoregulatory indoleamine 2,3 dioxygenase pathway in immunotherapy.
|
| |
Immunotherapy,
1,
645-661.
|
 |
|
|
|
|
 |
C.J.Austin,
F.Astelbauer,
P.Kosim-Satyaputra,
H.J.Ball,
R.D.Willows,
J.F.Jamie,
and
N.H.Hunt
(2009).
Mouse and human indoleamine 2,3-dioxygenase display some distinct biochemical and structural properties.
|
| |
Amino Acids,
36,
99.
|
 |
|
|
|
|
 |
E.Nickel,
K.Nienhaus,
C.Lu,
S.R.Yeh,
and
G.U.Nienhaus
(2009).
Ligand and substrate migration in human indoleamine 2,3-dioxygenase.
|
| |
J Biol Chem,
284,
31548-31554.
|
 |
|
|
|
|
 |
S.Löb,
A.Königsrainer,
H.G.Rammensee,
G.Opelz,
and
P.Terness
(2009).
Inhibitors of indoleamine-2,3-dioxygenase for cancer therapy: can we see the wood for the trees?
|
| |
Nat Rev Cancer,
9,
445-452.
|
 |
|
|
|
|
 |
A.Sheoran,
A.King,
A.Velasco,
J.M.Pero,
and
S.Garneau-Tsodikova
(2008).
Characterization of TioF, a tryptophan 2,3-dioxygenase involved in 3-hydroxyquinaldic acid formation during thiocoraline biosynthesis.
|
| |
Mol Biosyst,
4,
622-628.
|
 |
|
|
|
|
 |
J.B.Katz,
A.J.Muller,
and
G.C.Prendergast
(2008).
Indoleamine 2,3-dioxygenase in T-cell tolerance and tumoral immune escape.
|
| |
Immunol Rev,
222,
206-221.
|
 |
|
|
|
|
 |
J.L.Wee,
D.Christiansen,
Y.Q.Li,
W.Boyle,
and
M.S.Sandrin
(2008).
Suppression of cytotoxic and proliferative xenogeneic T-cell responses by transgenic expression of indoleamine 2,3-dioxygenase.
|
| |
Immunol Cell Biol,
86,
460-465.
|
 |
|
|
|
|
 |
J.Weigelt,
L.D.McBroom-Cerajewski,
M.Schapira,
Y.Zhao,
C.H.Arrowsmith,
and
C.H.Arrowmsmith
(2008).
Structural genomics and drug discovery: all in the family.
|
| |
Curr Opin Chem Biol,
12,
32-39.
|
 |
|
|
|
|
 |
M.von Grotthuss,
D.Plewczynski,
G.Vriend,
and
L.Rychlewski
(2008).
3D-Fun: predicting enzyme function from structure.
|
| |
Nucleic Acids Res,
36,
W303-W307.
|
 |
|
|
|
|
 |
S.J.Thackray,
C.G.Mowat,
and
S.K.Chapman
(2008).
Exploring the mechanism of tryptophan 2,3-dioxygenase.
|
| |
Biochem Soc Trans,
36,
1120-1123.
|
 |
|
|
|
|
 |
T.D.Bugg,
and
S.Ramaswamy
(2008).
Non-heme iron-dependent dioxygenases: unravelling catalytic mechanisms for complex enzymatic oxidations.
|
| |
Curr Opin Chem Biol,
12,
134-140.
|
 |
|
|
|
|
 |
E.R.Werner,
and
G.Werner-Felmayer
(2007).
Substrate and cofactor requirements of indoleamine 2,3-dioxygenase in interferon-gamma-treated cells: utilization of oxygen rather than superoxide.
|
| |
Curr Drug Metab,
8,
201-203.
|
 |
|
|
|
|
 |
F.Forouhar,
J.L.Anderson,
C.G.Mowat,
S.M.Vorobiev,
A.Hussain,
M.Abashidze,
C.Bruckmann,
S.J.Thackray,
J.Seetharaman,
T.Tucker,
R.Xiao,
L.C.Ma,
L.Zhao,
T.B.Acton,
G.T.Montelione,
S.K.Chapman,
and
L.Tong
(2007).
Molecular insights into substrate recognition and catalysis by tryptophan 2,3-dioxygenase.
|
| |
Proc Natl Acad Sci U S A,
104,
473-478.
|
 |
|
PDB codes:
|
 |
|
|
|
|
|
 |
A.J.Muller,
and
P.A.Scherle
(2006).
Targeting the mechanisms of tumoral immune tolerance with small-molecule inhibitors.
|
| |
Nat Rev Cancer,
6,
613-625.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
Where a reference describes a PDB structure, the PDB
codes are
shown on the right.
|
 |

