 |
PDBsum entry 2cd7

|

|

|
|
 |
Contents |
 |
|
|

|

|
|
|
|
|
|
|
|
* Residue conservation analysis
|
|
|

|
|
PDB id:
|
 |
|
 |
| Name: |
 |
Hydrolase
|
 |
|
Title:
|
 |
Staphylococcus aureus pi258 arsenate reductase (arsc) h62q mutant
|
|
Structure:
|
 |
Protein arsc. Chain: a. Synonym: arsenate reductase, arsenical pump modifier, low molecular weight protein-tyrosine-phosphatase. Engineered: yes. Mutation: yes. Other_details: reduced
|
|
Source:
|
 |
Staphylococcus aureus. Organism_taxid: 1280. Expressed in: escherichia coli. Expression_system_taxid: 511693.
|
|
Resolution:
|
 |
|
1.50Å
|
R-factor:
|
0.185
|
R-free:
|
0.218
|
|
|
Authors:
|
 |
L.Buts,G.Roos,K.Van Belle,E.Brosens,R.Loris,L.Wyns,J.Messens
|
Key ref:

|
 |
G.Roos
et al.
(2006).
Interplay between ion binding and catalysis in the thioredoxin-coupled arsenate reductase family.
J Mol Biol,
360,
826-838.
PubMed id:
DOI:

|
 |
|
Date:
|
 |
|
23-Jan-06
|
Release date:
|
28-Jun-06
|
|
|

|

|
|
PROCHECK
|

|

|
|
|
Headers
|
 |
|
|
References
|
|
|
|

|

|
|
P0A006
(ARSC_STAAU) -
Arsenate reductase from Staphylococcus aureus
|
|

|

|
Seq:
Struc:
|
 |
 |
 |
131 a.a.
131 a.a.*
|

|

|

|

|
|

|

|

|
 |
 |

|
|
Key: |
 |
PfamA domain |
 |
 |
 |
Secondary structure |
 |
 |
CATH domain |
 |
|
*
PDB and UniProt seqs differ
at 1 residue position (black
cross)
|
|

|
|
|
 |
|
|
 |
 |
 |
 |
Enzyme class:
|
 |
E.C.1.20.4.4
- arsenate reductase (thioredoxin).
|
|
 |
 |
 |
 |
 |
Reaction:
|
 |
arsenate + [thioredoxin]-dithiol + H+ = arsenite + [thioredoxin]- disulfide + H2O
|
 |
 |
 |
 |
 |
 arsenate
arsenate
|
+
|
[thioredoxin]-dithiol
|
+
|
H(+)
|
=
|
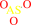 arsenite
arsenite
|
+
|
[thioredoxin]- disulfide
|
+
|
H2O
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Molecule diagrams generated from .mol files obtained from the
KEGG ftp site
|
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |
 |

|

|

|
| |
|
|
| |
|
DOI no:
|
J Mol Biol
360:826-838
(2006)
|
|
PubMed id:
|
|
|
|

|
| |
|
Interplay between ion binding and catalysis in the thioredoxin-coupled arsenate reductase family.
|
|
G.Roos,
L.Buts,
K.Van Belle,
E.Brosens,
P.Geerlings,
R.Loris,
L.Wyns,
J.Messens.
|
|
|

|
| |
ABSTRACT
|
|

|
| |

|
In the thioredoxin (Trx)-coupled arsenate reductase family, arsenate reductase
from Staphylococcus aureus plasmid pI258 (Sa_ArsC) and from Bacillus subtilis
(Bs_ArsC) are structurally related detoxification enzymes. Catalysis of the
reduction of arsenate to arsenite involves a P-loop
(Cys10Thr11Gly12Asn13Ser14Cys15Arg16) structural motif and a disulphide cascade
between three conserved cysteine residues (Cys10, Cys82 and Cys89). For its
activity, Sa_ArsC benefits from the binding of tetrahedral oxyanions in the
P-loop active site and from the binding of potassium in a specific
cation-binding site. In contrast, the steady-state kinetic parameters of Bs_ArsC
are not affected by sulphate or potassium. The commonly occurring mutation of a
histidine (H62), located about 6 A from the potassium-binding site in Sa_ArsC,
to a glutamine uncouples the kinetic dependency on potassium. In addition, the
binding affinity for potassium is affected by the presence of a lysine (K33) or
an aspartic acid (D33) in combination with two negative charges (D30 and E31) on
the surface of Trx-coupled arsenate reductases. In the P-loop of the Trx-coupled
arsenate reductase family, the peptide bond between Gly12 and Asn13 can adopt
two distinct conformations. The unique geometry of the P-loop with Asn13 in beta
conformation, which is not observed in structurally related LMW PTPases, is
stabilized by tetrahedral oxyanions and decreases the pK(a) value of Cys10 and
Cys82. Tetrahedral oxyanions stabilize the P-loop in its catalytically most
active form, which might explain the observed increase in k(cat) value for
Sa_ArsC. Therefore, a subtle interplay of potassium and sulphate dictates the
kinetics of Trx-coupled arsenate reductases.

|

|
|
|

|
| |
Selected figure(s)
|
|

|
| |
 |
 |

|
 |

|
 |
Figure 2.
Figure 2. Stereo view of the refined structure of Bs_ArsC.
Lysine 33 (blue), aspartate 30 and glutamate 31 (red), the
P-loop active site (residues 10–16) (red tube) with asparagine
13 (red stick representation), the redox-active cysteine
residues (yellow), sulphate (atom type), the cation-binding site
residues (green) and sodium (magenta) are shown. The Figure was
generated by using MacPyMol (Delano Scientific LLC 2005).
|
 |
Figure 6.
Figure 6. The conformational change in the P-loop of Sa_ArsC
(a) The 2F[o]–F[c] electron density map contoured at 1σ of
the P-loop of Sa_ArsC C10SC15A (PDB code 2FXI). (b) The P-loop
active site of Sa_ArsC C10SC15A harbouring a sulphate molecule
(PDB code 2FXI) (green) with the peptide bond between Gly12 and
Asn13 in a left-handed α[L] conformation. On top is the P-loop
(PDB code 1JFV)^8 (salmon) that binds a perchlorate (not shown)
with the peptide bond between Gly12 and Asn13 flipped to a β
conformation is visualized. Structures were superposed with the
SSM algorithm.^21 Figures were generated by using MacPyMol
(Delano Scientific LLC 2005).
|
 |
|
|

|
| |
The above figures are
reprinted
by permission from Elsevier:
J Mol Biol
(2006,
360,
826-838)
copyright 2006.
|
|
| |
Figures were
selected
by an automated process.
|
|

|

|
|
 |
 |
|
 |
 |
 |
 |
 |
 |
 |
 |
 |
|
Literature references that cite this PDB file's key reference
|
|
 |
| |
PubMed id
|
 |
Reference
|
 |
|
|
|
 |
G.Roos,
N.Foloppe,
K.Van Laer,
L.Wyns,
L.Nilsson,
P.Geerlings,
and
J.Messens
(2009).
How thioredoxin dissociates its mixed disulfide.
|
| |
PLoS Comput Biol,
5,
e1000461.
|
 |
|
|
|
|
 |
L.López-Maury,
A.M.Sánchez-Riego,
J.C.Reyes,
and
F.J.Florencio
(2009).
The glutathione/glutaredoxin system is essential for arsenate reduction in Synechocystis sp. strain PCC 6803.
|
| |
J Bacteriol,
191,
3534-3543.
|
 |
|
|
|
|
 |
O.Okhrimenko,
and
I.Jelesarov
(2008).
A survey of the year 2006 literature on applications of isothermal titration calorimetry.
|
| |
J Mol Recognit,
21,
1.
|
 |
|
 |
 |
|
The most recent references are shown first.
Citation data come partly from CiteXplore and partly
from an automated harvesting procedure. Note that this is likely to be
only a partial list as not all journals are covered by
either method. However, we are continually building up the citation data
so more and more references will be included with time.
|
 |

